6.2: Eukaryotic Gene Regulation
- Page ID
- 25749
Learning Objectives
- Describe the cis-acting elements that control tissue-specific transcription of eukaryotic protein encoding genes: promoters, enhancers, insulators and how trans-acting transcription factors can influence gene expression.
- Predict the effects of mutations in regulatory sequences on gene expression.
Like prokaryotes, transcriptional regulation in eukaryotes involves both cis-elements and trans-factors; however, there are more factors and they interact in more complex ways, many of which are still be elucidated.
Regulatory sequences
Proximal regulatory sequences
As in prokaryotes the RNA polymerase binds to the gene at its promoter to begin transcription. In eukaryotes, however, RNA polymerase is part of a large protein complex that includes additional proteins (transcription initiation factors) that bind to one or more specific cis-elements in the promoter region, including GC-rich boxes, CAAT boxes, and TATA boxes. High levels of transcription require both the presence of this protein complex at the promoter, as well as their interaction with other trans-factors described below. The approximate position of these elements is described relative to the transcription start site (+1), but the distance between any of these elements and the transcription start site can vary. These sites are typically within ~200 base pairs of the start of transcription.
Distal regulatory elements
Even more variation is observed in the position and orientation of the second major type of cis-regulatory element in eukaryotes, which are DNA sequences called enhancer elements or simply enhancers. Regulatory trans-factor proteins called transcription factors bind to specific enhancer sequences, then, while still bound to DNA, these proteins interact with RNA polymerase and other proteins at the promoter to enhance the rate of transcription. There are a wide variety of different transcription factors and each recognizes a specific DNA sequence (enhancer element) to promote gene expression in the adjacent gene under specific circumstances. Enhancers can be located near (~100s of bp) or far (~10K of bp), and either upstream or downstream, from the promoter. The transcription factors and the proteins associated with transcription initiation complex will bind each other, bending the DNA in the process.
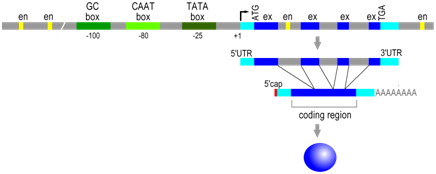
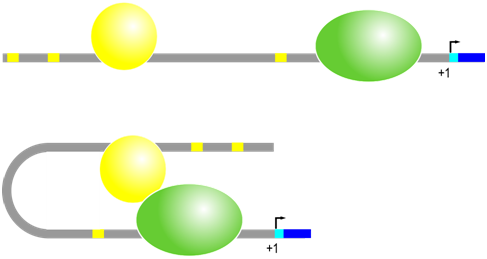
Enhancer sequence example
Some genes are transcriptionally regulated in response to hormones, such as estrogen. Estrogen receptors are proteins that bind the hormone estrogen and the the protein dimers are transported into the nucleus where they recognize sequences called estrogen response elements (EREs). The consensus ERE sequence is 5′-GGTCAnnnTGACC-3′ (where n is any base) at many locations in a genome and recruit RNA polymerase to induce the transcription of the corresponding genes. This consensus sequence was elucidated by studies of a human cell line to which a Xenopus (frog) estrogen responsive gene had been added, revealing that the roles of estrogen receptors in binding EREs and activating transcription of target genes is conserved across species.
Klein-Hitpass L, Schorpp M, Wagner U, Ryffel GU. An estrogen-responsive element derived from the 5' flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986 Sep 26;46(7):1053-61. doi: 10.1016/0092-8674(86)90705-1. PMID: 3463433.
Reviewed by Klinge C. Nucleic Acids Res. 2001 Jul 15; 29(14): 2905–2919.
Enhancers regulate the Drosophila yellow gene
The yellow gene of Drosophila provides an example of the modular nature of enhancers. This gene encodes an enzyme in the pathway that produces a dark pigment in the insect exoskeleton. Mutants have a yellow cuticle rather than the wild type darker pigmented cuticle. (Why call the gene “yellow”? Recall that genes are often named after their mutant phenotype.) Three enhancer elements each bind a different tissue-specific transcription factor in the wing, body, or mouth to enhance transcription of yellow in that tissue and make the dark pigment. So, the wing cells will have a transcription factor that binds to the wing enhancer to drive expression; likewise in the body and mouth cells. Thus, specific combinations of cis-elements and trans-factors control the differential, tissue-specific expression of genes. This type of combinatorial action of enhancers is typical of the transcriptional activation of most eukaryotic genes: specific transcription factors activate the transcription of target genes under specific conditions.
While enhancer sequences promote expression, there is an oppositely acting type of element, called silencers. These elements function in much the same manner, with transcription factors that bind to DNA sequences, but they act to repress or reduce transcription from the adjacent gene. A gene’s complete expression profile (transcription level, tissue specific, temporal specific) is a combination of various enhancer and silencer elements

Thinking about regulation of transcription factors
Understanding the transcription factors that regulate the expression of the yellow gene can prompt us to consider: why are those transcription factors present in different tissues? These transcription factors themselves are regulated by their own sets of transcription factors and so on and so on. Where does it begin? For Drosophila, the answer is in the egg! Specific proteins and mRNAs in the egg define the regions of the future embryo and a cascade of gene expression events lead to each adult cell and tissue type expressing the correct genes for their function.
Exercise \(\PageIndex{1}\)
What is the phenotype of the fruit fly when the yellow gene and enhancers are fully functional?
What would be the effect of a loss-of-function mutation in the yellow coding sequence?
What would be the effect of a mutation in the wing enhancer?
What would be the effect of a mutation in the TATA box region?
- Answer
-
The wild-type fruit fly has a dark black/brown cuticle.
A loss-of-function mutation in the yellow gene would lead to flies with yellow appearance, instead of brown/black.
A mutation in the wing enhancer would result in yellow wings, but other parts of cuticle should be unaffected.
A mutation in the TATA box should result in a phenotype similar to loss-of-function in the yellow gene.
Contributors and Attributions
Dr. Todd Nickle and Isabelle Barrette-Ng (Mount Royal University) The content on this page is licensed under CC SA 3.0 licensing guidelines.
Using reporter genes to study transcription


