12.2: Antigens, Antigen Presenting Cells, and Major Histocompatibility Complexes
- Page ID
- 23670
Skills to Develop
- Identify cells that express MHC I and/or MHC II molecules and describe the structures and cellular location of MHC I and MHC II molecules
- Identify the cells that are antigen-presenting cells
- Describe the process of antigen processing and presentation with MHC I and MHC II
As discussed in Cellular Defenses, major histocompatibility complex (MHC) molecules are expressed on the surface of healthy cells, identifying them as normal and “self” to natural killer (NK) cells. MHC molecules also play an important role in the presentation of foreign antigens, which is a critical step in the activation of T cells and thus an important mechanism of the adaptive immune system.
Major Histocompatibility Complex Molecules
The major histocompatibility complex (MHC) is a collection of genes coding for MHC molecules found on the surface of all nucleated cells of the body. In humans, the MHC genes are also referred to as human leukocyte antigen (HLA) genes. Mature red blood cells, which lack a nucleus, are the only cells that do not express MHC molecules on their surface.
There are two classes of MHC molecules involved in adaptive immunity, MHC I and MHC II (Figure \(\PageIndex{1}\)). MHC I molecules are found on all nucleated cells; they present normal self-antigens as well as abnormal or nonself pathogens to the effector T cells involved in cellular immunity. In contrast, MHC II molecules are only found on macrophages, dendritic cells, and B cells; they present abnormal or nonself pathogen antigens for the initial activation of T cells.
Both types of MHC molecules are transmembrane glycoproteins that assemble as dimers in the cytoplasmic membrane of cells, but their structures are quite different. MHC I molecules are composed of a longer α protein chain coupled with a smaller β2 microglobulin protein, and only the α chain spans the cytoplasmic membrane. The α chain of the MHC I molecule folds into three separate domains: α1, α2 and α3. MHC II molecules are composed of two protein chains (an α and a β chain) that are approximately similar in length. Both chains of the MHC II molecule possess portions that span the plasma membrane, and each chain folds into two separate domains: α1 and α2, and β1, and β2. In order to present abnormal or non-self-antigens to T cells, MHC molecules have a cleft that serves as the antigen-binding site near the “top” (or outermost) portion of the MHC-I or MHC-II dimer. For MHC I, the antigen-binding cleft is formed by the α1 and α2 domains, whereas for MHC II, the cleft is formed by the α1 and β1 domains (Figure \(\PageIndex{1}\)).
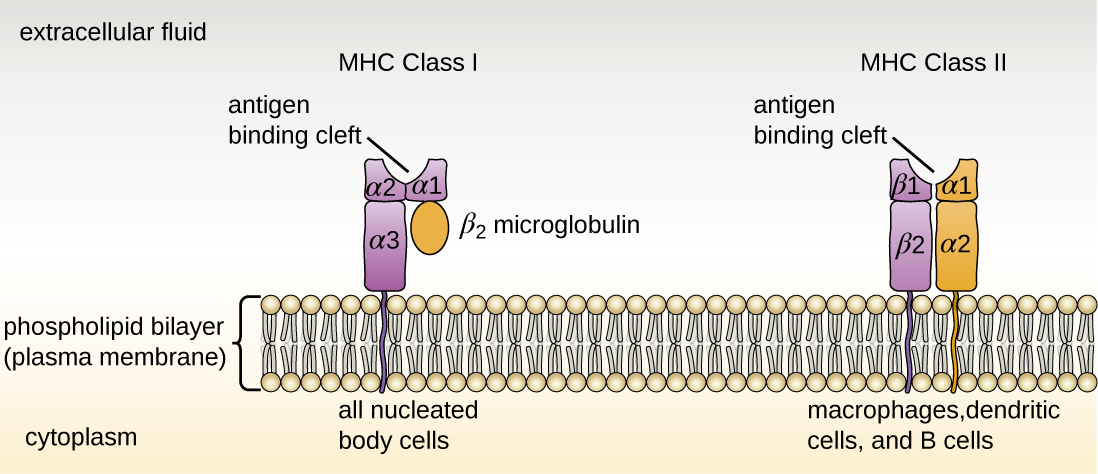
Figure \(\PageIndex{1}\): MHC I are found on all nucleated body cells, and MHC II are found on macrophages, dendritic cells, and B cells (along with MHC I). The antigen-binding cleft of MHC I is formed by domains α1 and α2. The antigen-binding cleft of MHC II is formed by domains α1 and β1.
Exercise \(\PageIndex{1}\)
Compare the structures of the MHC I and MHC II molecules.
Antigen-Presenting Cells (APCs)
All nucleated cells in the body have mechanisms for processing and presenting antigens in association with MHC molecules. This signals the immune system, indicating whether the cell is normal and healthy or infected with an intracellular pathogen. However, only macrophages, dendritic cells, and B cells have the ability to present antigens specifically for the purpose of activating T cells; for this reason, these types of cells are sometimes referred to as antigen-presenting cells (APCs).
While all APCs play a similar role in adaptive immunity, there are some important differences to consider. Macrophages and dendritic cells are phagocytes that ingest and kill pathogens that penetrate the first-line barriers (i.e., skin and mucous membranes). B cells, on the other hand, do not function as phagocytes but play a primary role in the production and secretion of antibodies. In addition, whereas macrophages and dendritic cells recognize pathogens through nonspecific receptor interactions (e.g., PAMPs, toll-like receptors, and receptors for opsonizing complement or antibody), B cells interact with foreign pathogens or their free antigens using antigen-specific immunoglobulin as receptors (monomeric IgD and IgM). When the immunoglobulin receptors bind to an antigen, the B cell internalizes the antigen by endocytosis before processing and presentting the antigen to T cells.
Antigen Presentation with MHC II Molecules
MHC II molecules are only found on the surface of APCs. Macrophages and dendritic cells use similar mechanisms for processing and presentation of antigens and their epitopes in association with MHC II; B cells use somewhat different mechanisms that will be described further in B Lymphocytes and Humoral Immunity. For now, we will focus on the steps of the process as they pertain to dendritic cells.
After a dendritic cell recognizes and attaches to a pathogen cell, the pathogen is internalized by phagocytosis and is initially contained within a phagosome. Lysosomes containing antimicrobial enzymes and chemicals fuse with the phagosome to create a phagolysosome, where degradation of the pathogen for antigen processing begins. Proteases (protein-degrading) are especially important in antigen processing because only protein antigen epitopes are presented to T cells by MHC II (Figure \(\PageIndex{2}\)).
APCs do not present all possible epitopes to T cells; only a selection of the most antigenic or immunodominantepitopes are presented. The mechanism by which epitopes are selected for processing and presentation by an APC is complicated and not well understood; however, once the most antigenic, immunodominant epitopes have been processed, they associate within the antigen-binding cleft of MHC II molecules and are translocated to the cell surface of the dendritic cell for presentation to T cells.
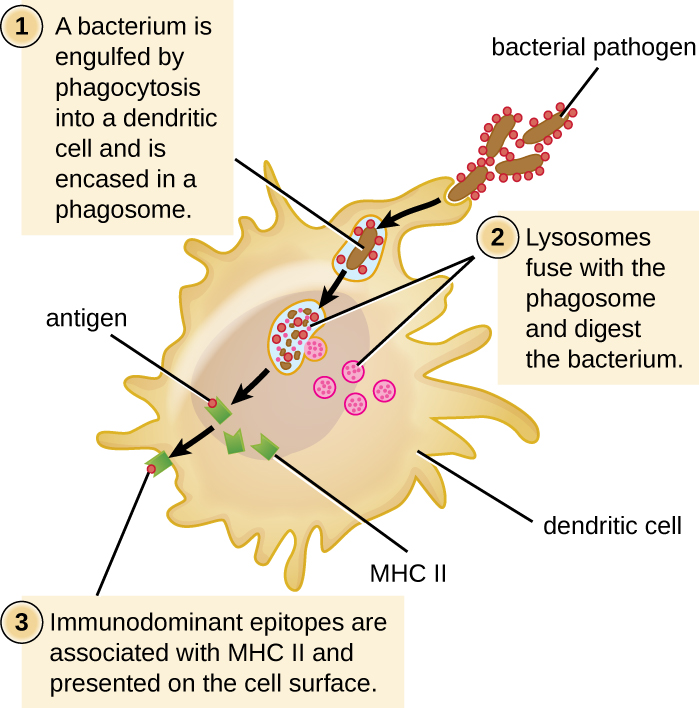
Figure \(\PageIndex{2}\): A dendritic cell phagocytoses a bacterial cell and brings it into a phagosome. Lysosomes fuse with the phagosome to create a phagolysosome, where antimicrobial chemicals and enzymes degrade the bacterial cell. Proteases process bacterial antigens, and the most antigenic epitopes are selected and presented on the cell’s surface in conjunction with MHC II molecules. T cells recognize the presented antigens and are thus activated.
Exercise \(\PageIndex{2}\)
- What are the three kinds of APCs?
- What role to MHC II molecules play in antigen presentation?
- What is the role of antigen presentation in adaptive immunity?
Antigen Presentation with MHC I Molecules
MHC I molecules, found on all normal, healthy, nucleated cells, signal to the immune system that the cell is a normal “self” cell. In a healthy cell, proteins normally found in the cytoplasm are degraded by proteasomes (enzyme complexes responsible for degradation and processing of proteins) and processed into self-antigen epitopes; these self-antigen epitopes bind within the MHC I antigen-binding cleft and are then presented on the cell surface. Immune cells, such as NK cells, recognize these self-antigens and do not target the cell for destruction. However, if a cell becomes infected with an intracellular pathogen (e.g., a virus), protein antigens specific to the pathogen are processed in the proteasomes and bind with MHC I molecules for presentation on the cell surface. This presentation of pathogen-specific antigens with MHC I signals that the infected cell must be targeted for destruction along with the pathogen.
Before elimination of infected cells can begin, APCs must first activate the T cells involved in cellular immunity. If an intracellular pathogen directly infects the cytoplasm of an APC, then the processing and presentation of antigens can occur as described (in proteasomes and on the cell surface with MHC I). However, if the intracellular pathogen does not directly infect APCs, an alternative strategy called cross-presentation is utilized. In cross-presentation, antigens are brought into the APC by mechanisms normally leading to presentation with MHC II (i.e., through phagocytosis), but the antigen is presented on an MHC I molecule for CD8 T cells. The exact mechanisms by which cross-presentation occur are not yet well understood, but it appears that cross-presentation is primarily a function of dendritic cells and not macrophages or B cells.
Exercise \(\PageIndex{3}\)
- Compare and contrast antigen processing and presentation associated with MHC I and MHC II molecules.
- What is cross-presentation, and when is it likely to occur?
Key Concepts and Summary
- Major histocompatibility complex (MHC) is a collection of genes coding for glycoprotein molecules expressed on the surface of all nucleated cells.
- MHC I molecules are expressed on all nucleated cells and are essential for presentation of normal “self” antigens. Cells that become infected by intracellular pathogens can present foreign antigens on MHC I as well, marking the infected cell for destruction.
- MHC II molecules are expressed only on the surface of antigen-presenting cells (macrophages, dendritic cells, and B cells). Antigen presentation with MHC II is essential for the activation of T cells.
- Antigen-presenting cells (APCs) primarily ingest pathogens by phagocytosis, destroy them in the phagolysosomes, process the protein antigens, and select the most antigenic/immunodominant epitopes with MHC II for presentation to T cells.
- Cross-presentation is a mechanism of antigen presentation and T-cell activation used by dendritic cells not directly infected by the pathogen; it involves phagocytosis of the pathogen but presentation on MHC I rather than MHC II.
Multiple Choice
MHC I molecules present
A. processed foreign antigens from proteasomes.
B. processed self-antigens from phagolysosome.
C. antibodies.
D. T cell antigens.
A
MHC II molecules present
A. processed self-antigens from proteasomes.
B. processed foreign antigens from phagolysosomes.
C. antibodies.
D. T cell receptors.
B
Which type of antigen-presenting molecule is found on all nucleated cells?
A. MHC II
B. MHC I
C. antibodies
D. B-cell receptors
B
Which type of antigen-presenting molecule is found only on macrophages, dendritic cells, and B cells?
A. MHC I
B. MHC II
C. T-cell receptors
D. B-cell receptors
B
Fill in the Blank
MHC molecules are used for antigen ________ to T cells.
presentation
MHC II molecules are made up of two subunits (α and β) of approximately equal size, whereas MHC I molecules consist of a larger α subunit and a smaller subunit called ________.
β2 microglobulin
Critical Thinking
Which mechanism of antigen presentation would be used to present antigens from a cell infected with a virus?
Which pathway of antigen presentation would be used to present antigens from an extracellular bacterial infection?
Contributor
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)


