8.1: Introduction to Bacterial Identification using Culture Media
- Page ID
- 52313
Learning Outcomes
- Identify and describe culture media for the growth and identification of bacteria, including examples of selective and/or differential media.
Bacterial Identification with Culture media
One of the common methods to identify bacteria is through the use of specialized media. Back in Module 2, you were introduced to this idea. Here’s a recap: In clinical microbiology, it is often important to detect the presence of specific microbes associated with disease or poor sanitation, for this task, selective and/or differential media are used. Selective media suppresses the growth of unwanted bacteria and encourage the growth of desired microbes. For example, bismuth sulfite agar can be used to isolate Salmonella typhi from feces. Bismuth sulfite inhibits Gram positive bacteria and most Gram negative intestinal bacteria (other than S. typhi).
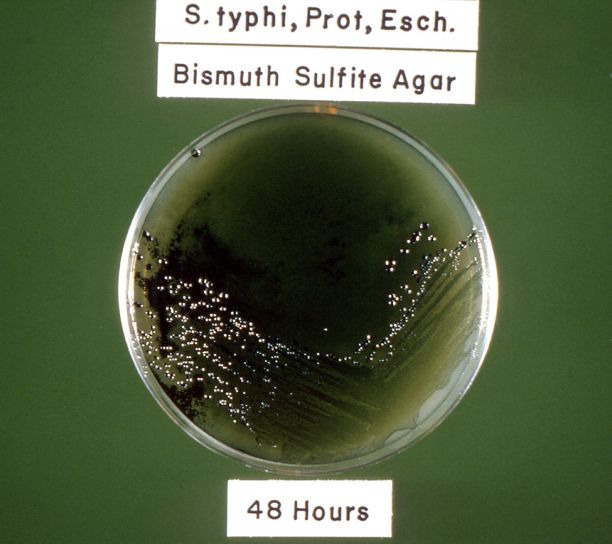
Image 1: Bismuth Sulfite agar
Differential media makes it easier to distinguish colonies of your desired microorganism from other colonies growing on the same plate. Blood agar (which contains red blood cells/RBCs) is a medium often used to identify bacterial species that destroy RBCs. These species, such as Streptococcus pyogenes, that causes strep throat, will show a clear ring around their colonies where they have lysed the surrounding blood cells.
Image 2: Normal Upper respiratory flora mixed with Streptococcus species. The presence of beta-hemolytic colonies (clear zones around small colonies) indicates the possibility of Streptococcus pyogenes infection. Image by Rebecca Buxton, University of Utah, Salt Lake City, UT.
Selective and Differential Media types
A. Mannitol Salt Agar
In some cases, selective and differential characteristics are combined into a single medium. If you wanted to isolate the common bacterium, Staphylococcus aureus and know that it has a tolerance for high concentrations of sodium chloride and can ferment the carbohydrate mannitol to form acids, you can use a selective-differential media called “Mannitol Salt Agar or MSA” which contains 7.5% sodium chloride, and will discourage the growth of competing microbes and thus select for (or favor the growth of) S. aureus. MSA is too salty for many microbes. The medium also contains the pH indicator, phenol red, when the pH drops, such as when mannitol in the medium is fermented to acids, it will change color from red to yellow. Thus, the mannitol-fermenting colonies of S. aureus are differentiated from colonies of bacteria that do not ferment mannitol. Bacteria that can grow at high salt concentration and ferment mannitol to acid can be identified on MSA by color change, such as shown on this image 3-S. aureus streak in the left half of the plate shows growth of the bacterium and the yellow color indicating mannitol fermentation, while on the right half of the plate, Streptococcus durans is not capable of growing on this media (because it cannot tolerate high salt concentration) and thus there are no colonies and the media remains the same color, a pinkish red. If a bacterium is able to grow on MSA (has visible colonies) but the medium stays pinkish-red, then it is not capable of fermenting mannitol since the medium doesn't change color.
Image 3: Mannitol salt agar inoculated with Staphylococcus aureus on the left side of the plate and showing fermentation of mannitol (yellow medium) and inoculated with Streptococcus durans on the right side of the plate, which shows no growth (no colonies visible, medium remains reddish pink). Image by Anne Y. Tsang and Patricia Shields, University of Maryland, College Park, MD.
Watch Video 1: Mannitol Salt Agar explained
Watch Video 1: Mannitol Salt Agar explained and showing plate examples. Video by Dr. Gary Kaiser (CCCB ). (1:38) URL: https://youtu.be/kG1_Tf5Vpc0
B. MacConkey agar
MacConkey agar is another example of a Selective & differential medium, it is used to isolate & distinguish between Gram-negative enteric rods. MacConkey agar contains bile salts and crystal violet that inhibit Gram positive bacteria but allow the growth of Gram negative bacteria. In this way, these chemicals act selectively, to inhibit one group but promote the growth of another group.
MacConkey also has the sugar lactose and a pH indicator neutral red it turns pink/red under acid conditions. In this way, it distinguishes between lactose fermenters from non-lactose fermenters.
Image 4: MacConkey agar plate inoculated with the Gram-negative lactose fermenter (pink colonies/streak) Escherichia coli and the Gram-negative non-lactose fermenter (off yellow colonies) Serratia marcescens. Image by David Miller and Patrick Hanley, Hartwick College, Oneonta, NY)
Watch video 2: MacConkey agar explained
Watch video 2: MacConkey agar explained and showing plate examples. Video by Dr. Gary Kaiser (CCCB ). (4:32) URL: https://youtu.be/yInQ9jApAlU
C. Eosin-Methylene Blue (EMB) agar
Eosin-Methylene Blue (EMB) agar is a selective & differential medium used to isolate Gram-negative enteric bacteria and distinguish between them. EMB contains 2 dyes--Eosin and methylene blue--which act as selective agents to inhibit Gram-positive bacteria but allow for the growth of Gram-negative bacteria.
EMB also contains the sugars lactose and sucrose--and it distinguishes or differentiates between lactose or sucrose fermenters from non-fermenters. The fermenters form colored colonies and the non-fermenters form colorless colonies, similar to the MacConkey agar.
Image 5: Eosin-methylene blue ( EMB) agar plate inoculated with Escherichia coli (a Gram-negative coliform bacterium) showing good growth of dark blue-black colonies with metallic green sheen indicating vigorous fermentation of lactose and acid production which precipitates the green metallic pigment. Image by Naowarat Cheeptham, Thompson Rivers University, Kamloops, BC, Canada.
D. Hektoen Enteric (HE) agar
Hektoen Enteric (HE) agar is a selective & differential medium that is used to isolate and distinguish between the enteric pathogens Salmonella and Shigella. HE medium contains high concentrations of bile salts as selective agents that inhibit Gram-positive bacteria but also slow the growth of normal intestinal microbiota. HE contains sugars lactose and sucrose, the pH indicator bromothymol blue, sodium thiosulfate and ferric salts. E. coli and related coliforms ferment lactose (and sucrose) forming bright orange to salmon pink colonies. Salmonella and Shigella are non-lactose fermenters and form green or blue green colonies. Salmonella colonies produce Hydrogen sulfide (H2S) which reacts with the iron in the medium to form iron sulfide (FeS) forming colonies with black centers.
All of the media we discussed are for isolating and differentiating common enteric pathogens that can cause disease in humans.
Image 6: Hektoen enteric agar sterile, uninoculated
Image 7: Salmonella enterica on Hektoen enteric agar. Note the black center of the transparent colonies, indicating H2S production in the absence of carbohydrate fermentation. All serotypes of Salmonella have this appearance on Hektoen enteric agar except for serotype typhi, which is a weak H2S producer. Image by Jan Hudzicki, University of Kansas Medical Center, Kansas City, KS
Image 8: Enterobacter aerogenes on Hektoen enteric agar. Note the yellow-orange colonies, indicating the fermentation of at least one of the carbohydrates present in the medium. The lack of black colonies indicates no H2S production. The orange haze around the colonies is due to the precipitation of the bile salts by the organism. The appearance of E. aerogenes on Hektoen enteric agar is typical of most nonpathogenic enteric Gram-negative rods. Image by Jan Hudzicki, University of Kansas Medical Center, Kansas City, KS.

