3.2: Viral Evolution
- Page ID
- 80167
Natural Selection
Natural selection is the process underlying our understanding of evolution. Although viruses are not alive, they can evolve. Natural selection relies on four principles which can be applied to viruses:
- characteristics can be inherited
- For viruses, the genetic material that encodes heritable characteristics can be either DNA or RNA. For reasons explained below, RNA viruses tend to evolve more quickly than DNA viruses.
- more offspring are produced than are able to survive
- Each virus-infected cell can produce millions of viral particles, only a small fraction of which will successfully bind to and infect another cell.
- there is variation of characteristics within a population
- Genetic differences between viruses in a population can come about in three different ways (drift, shift, and recombination) which are explained below.
- offspring with more favorable characteristics reproduce more successfully
- Viruses are more successful at reproducing if they are better at 1) finding a host and 2) binding to the host cells. For animals, a virus-infected host that is less sick will interact with more potential hosts. Also, some variations of the glycoprotein spikes on the virus will bind better to the host cell receptors. Therefore, the general progression of viral evolution is to more contagious but less virulent variants (or strains). A good example of this is the emergence of the Omicron variant of SARS-CoV2, which is extremely contagious but appears to be less lethal than the original variant.
Genetic Drift
RNA-dependent RNA polymerases (RdRPs), unlike DNA polymerase or cellular RNA polymerase, tend to be very "sloppy" when synthesizing RNA. Because of this, there is a high frequency of mutations in the genomes of RNA viruses. Although this might at first seem detrimental to the virus, it is actually evolutionary beneficial. Each infected cell produces millions of new viral particles, most of which are the same or have mutations which do not substantially affect the virus's function. Some may have mutations which don't allow the virus to replicate at all. But occasionally, one of the variants produced has a mutation which allows it to spread more effectively through a population due to better binding to host cell receptors or increased reproduction rate. Sometimes a mutation changes the glycoprotein spikes enough to allow the virus to infect a new species. Such accumulation of mutations over time is called genetic drift (Figure \(\PageIndex{7A}\)). If the glycoprotein spikes of the virus are affected, it is specifically referred to as antigenic drift.
Genetic Shift
Genetic drift is not the only way for a virus to change, however. The genome of influenza virus consists of 8 segments of (-)ssRNA. Genetic shift can only happen in viruses with a segmented genome. On occasion, one individual can become infected with more than one strain (variant) of influenza virus. When the two different strains of virus infect one cell, the RNA segments of both viral strains are produced. When the new viral particles are assembled the will receive a mixture of segments from each strain. The viruses produced through this reshuffling of genomes are different from either of the original infecting strains. This is called genetic (or antigenic) shift (Figure \(\PageIndex{7B}\)). The most dangerous new strains of influenza often emerge as a result of antigenic shift because, unlike strains produced through drift, human immune systems have not encountered similar strains of influenza previously. When the terms antigenic drift and antigenic shift are used (as opposed to genetic drift or shift), it refers specifically to changes in the structures of the viral surface proteins which are recognized by the immune system. Proteins and other molecules which are detected by the immune system are called antigens. For influenza, these antigens are HA and NA. Strains of flu are categorized and named based on the types of HA and NA they have, such as the H1N1 flu of 2009 (type 1 HA and type 1 NA).
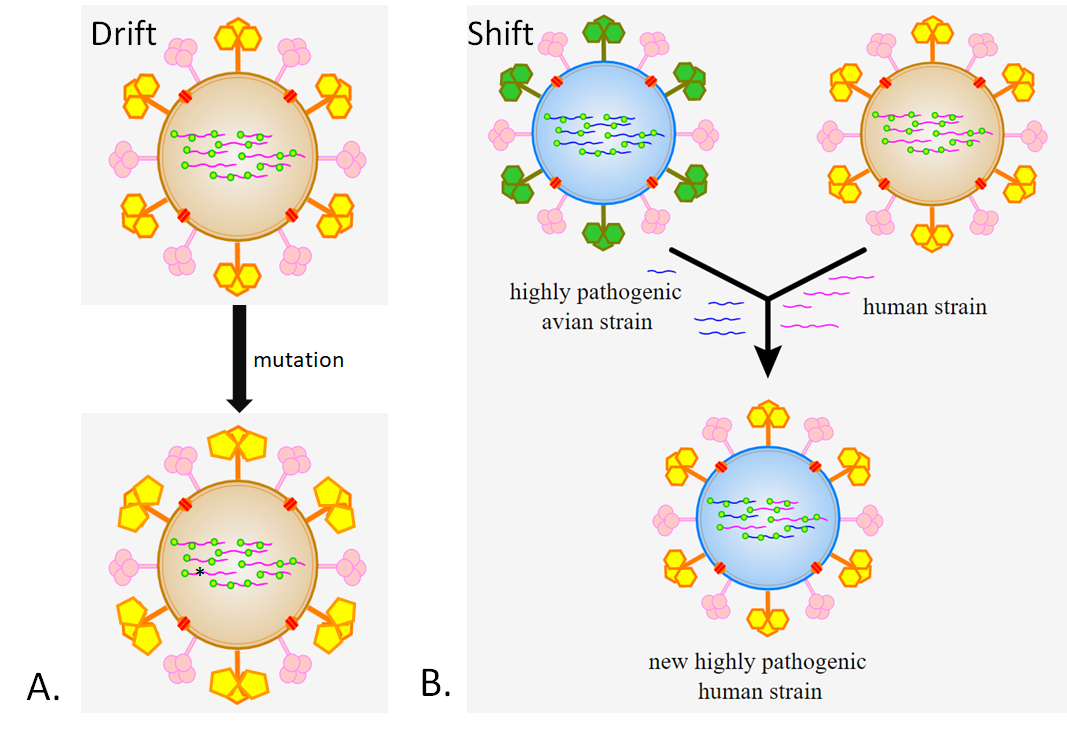
Figure \(\PageIndex{7}\): Genetic change in influenza virus through A) drift and B) shift. In drift, a slight change develops (represented by the yellow pentagons as opposed to hexagons on the glycoprotein spikes) a result of one or more mutations (represented by the * on the RNA segment). Shift results from the shuffling of the genomes of two different strains. Note the new strain has RNA from both parent strains (blue and pink). The glycoproteins (HA), however, are those of the human strain (yellow) allowing the new strain to bind to and infect human cells (modified from "File:Influenza geneticshift.svg" by Influenza_geneticshift.jpg: Dhorspool at en.wikipedia derivative work: Jiver is licensed under CC BY-SA 3.0)
Recombination
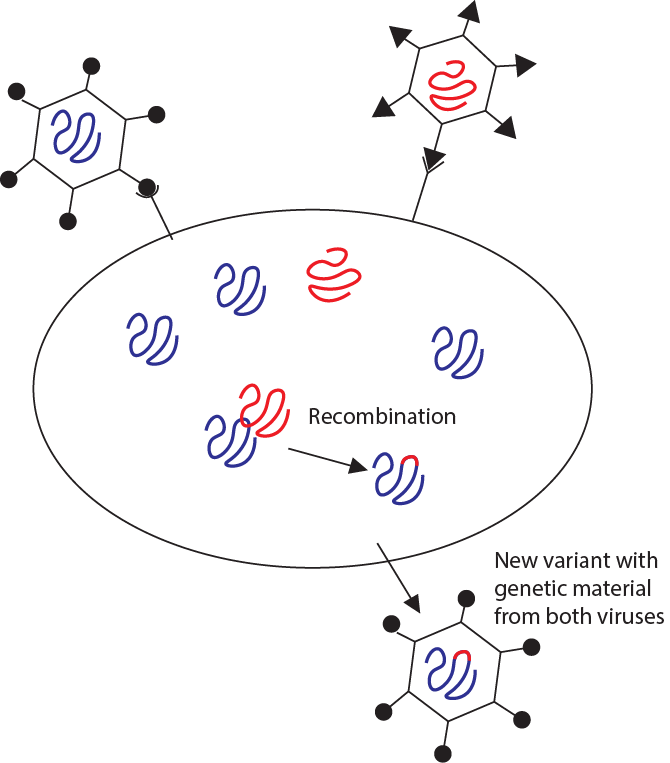
When genetic material (RNA or DNA) from different sources but with similar sequences encounter each other, they can sometimes undergo a process called recombination. (This frequently happens in eukaryotes during meiosis between homologous chromosomes.) In viruses, similar to genetic shift, recombination can happen if the same host cell is infected with two different but related viruses (Figure \(\PageIndex{8}\)). The new viral variants will carry mostly the genetic material from one virus, but will also carry a small portion of genetic material from the other. This phenomenon was observed in the Omicron variant of SARS-CoV2 which appears to carry some genes common in coronaviruses but not in other variants of SARS-CoV2.
This page contains content derived from OpenStax as well as new content authored by Jeanne Kagle.

