3.1.2: The Viral Life Cycle
- Page ID
- 42478
Learning Objectives
- Describe the lytic and lysogenic life cycles
- Describe the replication process of animal viruses
- Describe unique characteristics of retroviruses and latent viruses
- Discuss human viruses and their virus-host cell interactions
All viruses depend on cells for reproduction and metabolic processes. 3.1.2: The Viral Life CycleBy themselves, viruses do not encode for all of the enzymes necessary for viral replication. But within a host cell, a virus can commandeer cellular machinery to produce more viral particles. Bacteriophages replicate only in the cytoplasm, since prokaryotic cells do not have a nucleus or organelles. In eukaryotic cells, most DNA viruses can replicate inside the nucleus, with an exception observed in the large DNA viruses, such as the poxviruses, that can replicate in the cytoplasm. RNA viruses that infect animal cells often replicate in the cytoplasm.
General Viral Replication
All viruses follow the same six basic steps when replicating (Figure 3.2.1.1). Before a virus can do anything else it must 1) bind to a host cell (adhesion or attachment). This binding occurs between glycoprotein spikes on the surface of the viral particle and receptors on the surface of the host cell. This protein-protein binding is the primary factor in determining which cells a virus can infect. For instance, slight changes in the structure of the viral glycoprotein can allow a virus that normally infects another animal such as a bird or bat bind to human cell surface proteins, causing the virus to "jump" from animals to humans.
Once the virus is bound, 2) the genetic material enters the host cell. The genetic material can follow various paths based on the type of nucleic acid and the specific virus. Active replication, however requires 3) synthesis of viral proteins and 4) replication of the viral genome. Once both the proteins and genetic material are made, they are 5) assembled into viral particles. Finally, 6) the particles are released. If the virus is enveloped, the virus buds off the cell, taking a portion of the host cell membrane embedded with viral proteins (including the glycoprotein spikes for binding to host cells) with it. Otherwise, the host cell lyses, releasing the naked viral particles.
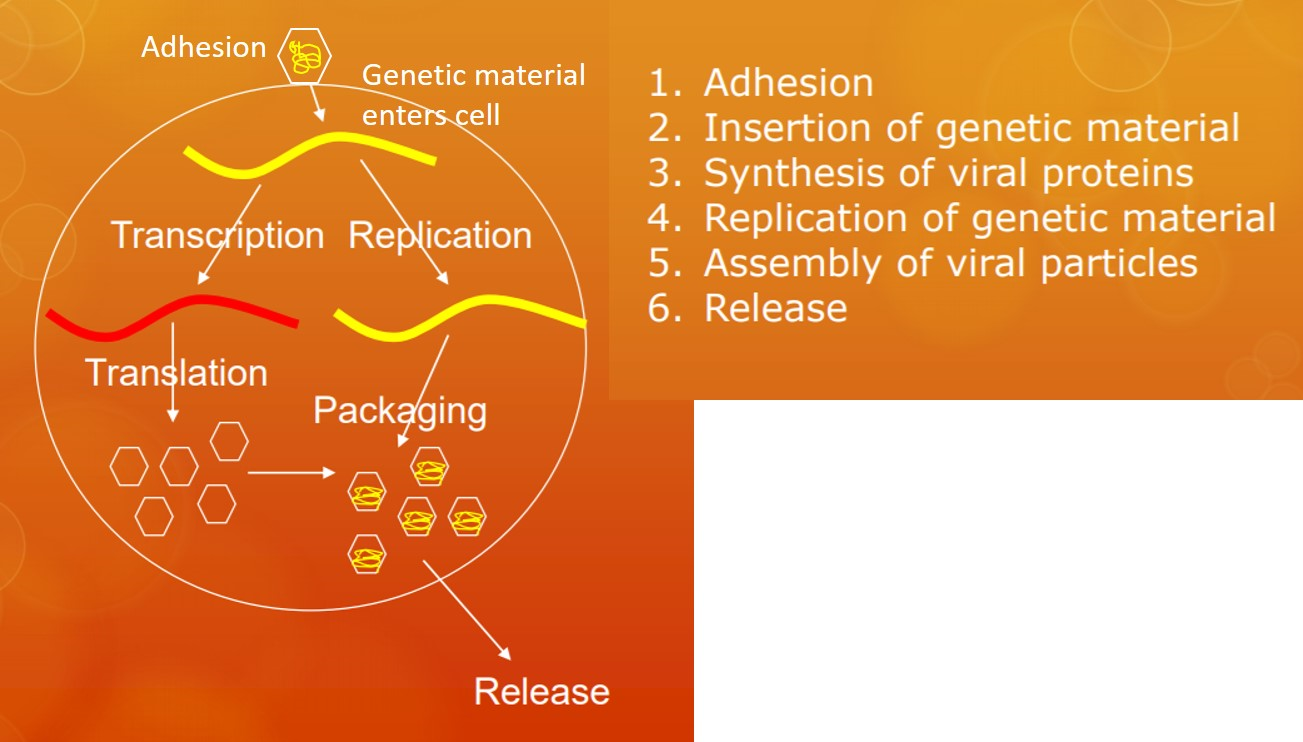
The Life Cycle of Viruses with Prokaryote Hosts
The life cycle of bacteriophages has been a good model for understanding how viruses affect the cells they infect, since similar processes have been observed for eukaryotic viruses, which can cause immediate death of the cell or establish a latent or chronic infection. Virulent phages typically lead to the death of the cell through cell lysis. Temperate phages, on the other hand, can become part of a host chromosome and are replicated with the cell genome until such time as they are induced to make newly assembled viruses, or progeny viruses.
The Lytic Cycle
During the lytic cycle of virulent phage, the bacteriophage takes over the cell, reproduces new phages, and destroys the cell. T-even phage is a good example of a well-characterized class of virulent phages. There are five stages in the bacteriophage lytic cycle (see Figure \(\PageIndex{2}\)). Attachment is the first stage in the infection process in which the phage interacts with specific bacterial surface receptors (e.g., lipopolysaccharides and OmpC protein (a porin) on host surfaces). Most phages have a narrow host range and may infect one species of bacteria or one strain within a species. This unique recognition can be exploited for targeted treatment of bacterial infection by phage therapy or for phage typing to identify unique bacterial subspecies or strains. The second stage of infection is entry or penetration. This occurs through contraction of the tail sheath, which acts like a hypodermic needle to inject the viral genome through the cell wall and membrane. The phage head and remaining components remain outside the bacteria.
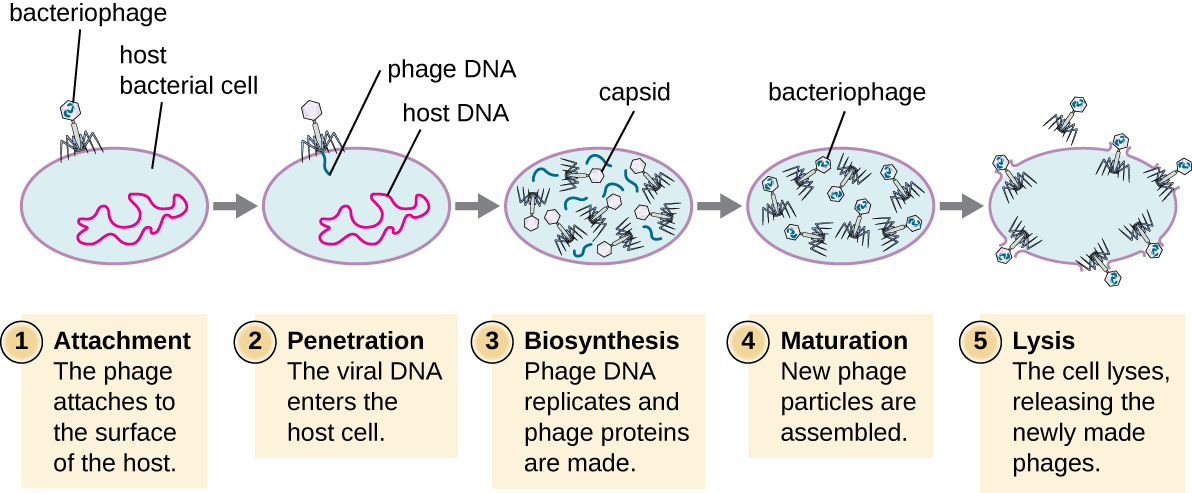
The third stage of infection is biosynthesis of new viral components. After entering the host cell, the virus synthesizes virus-encoded endonucleases to degrade the bacterial chromosome. It then hijacks the host cell to replicate, transcribe, and translate the necessary viral components (capsomeres, sheath, base plates, tail fibers, and viral enzymes) for the assembly of new viruses. Polymerase genes are usually expressed early in the cycle, while capsid and tail proteins are expressed later. During the maturation phase, new virions are created. To liberate free phages, the bacterial cell wall is disrupted by phage proteins such as holin or lysozyme. The final stage is release. Mature viruses burst out of the host cell in a process called lysis and the progeny viruses are liberated into the environment to infect new cells.
The Lysogenic Cycle
In a lysogenic cycle, the phage genome also enters the cell through attachment and penetration. A prime example of a phage with this type of life cycle is the lambda phage. During the lysogenic cycle, instead of killing the host, the phage genome integrates into the bacterial chromosome and becomes part of the host. The integrated phage genome is called a prophage. A bacterial host with a prophage is called a lysogen. The process in which a bacterium is infected by a temperate phage is called lysogeny. It is typical of temperate phages to be latent or inactive within the cell. As the bacterium replicates its chromosome, it also replicates the phage’s DNA and passes it on to new daughter cells during reproduction. The presence of the phage may alter the phenotype of the bacterium, since it can bring in extra genes (e.g., toxin genes that can increase bacterial virulence). This change in the host phenotype is called lysogenic conversion or phage conversion. Some bacteria, such as Vibrio cholerae and Clostridium botulinum, are less virulent in the absence of the prophage. The phages infecting these bacteria carry the toxin genes in their genome and enhance the virulence of the host when the toxin genes are expressed. In the case of V. cholera, phage encoded toxin can cause severe diarrhea; in C. botulinum, the toxin can cause paralysis. During lysogeny, the prophage will persist in the host chromosome until induction, which results in the excision of the viral genome from the host chromosome. After induction has occurred the temperate phage can proceed through a lytic cycle and then undergo lysogeny in a newly infected cell (see Figure \(\PageIndex{3}\)).
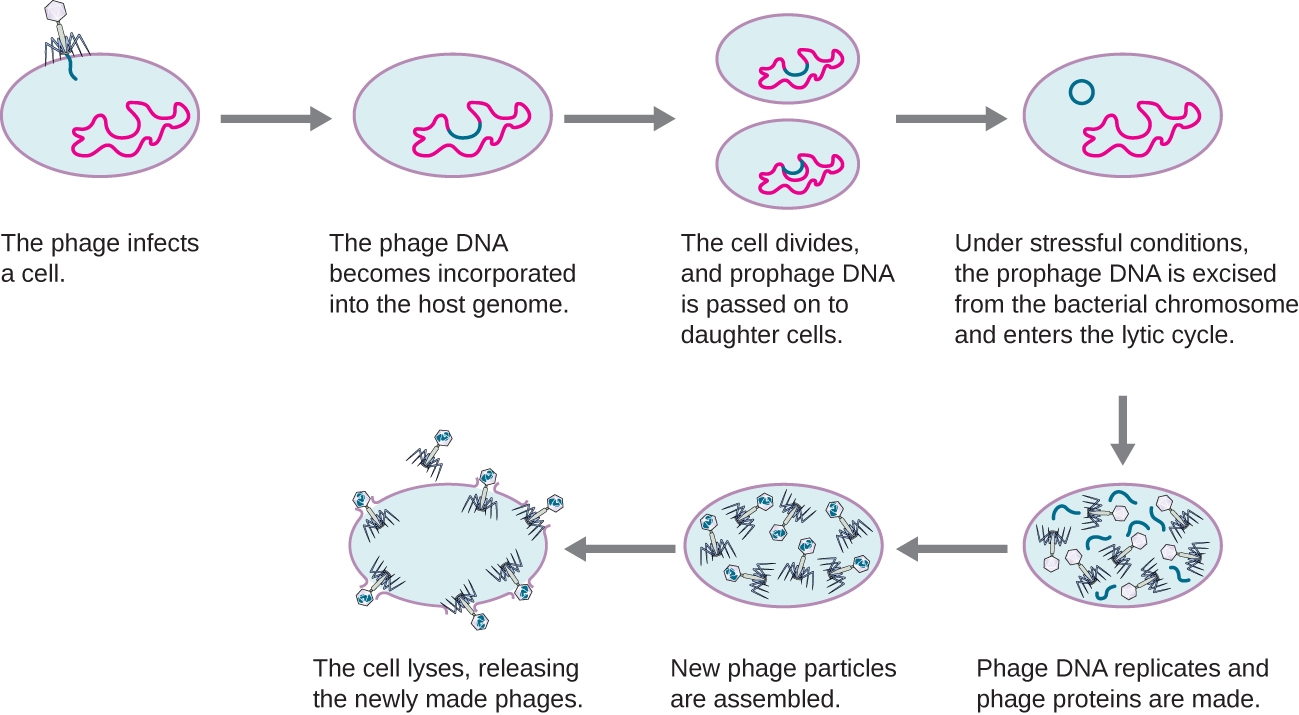
This video illustrates the stages of the lysogenic life cycle of a bacteriophage and the transition to a lytic phase.
Exercise \(\PageIndex{1}\)
Is a latent phage undetectable in a bacterium?
Life Cycle of Viruses with Animal Hosts
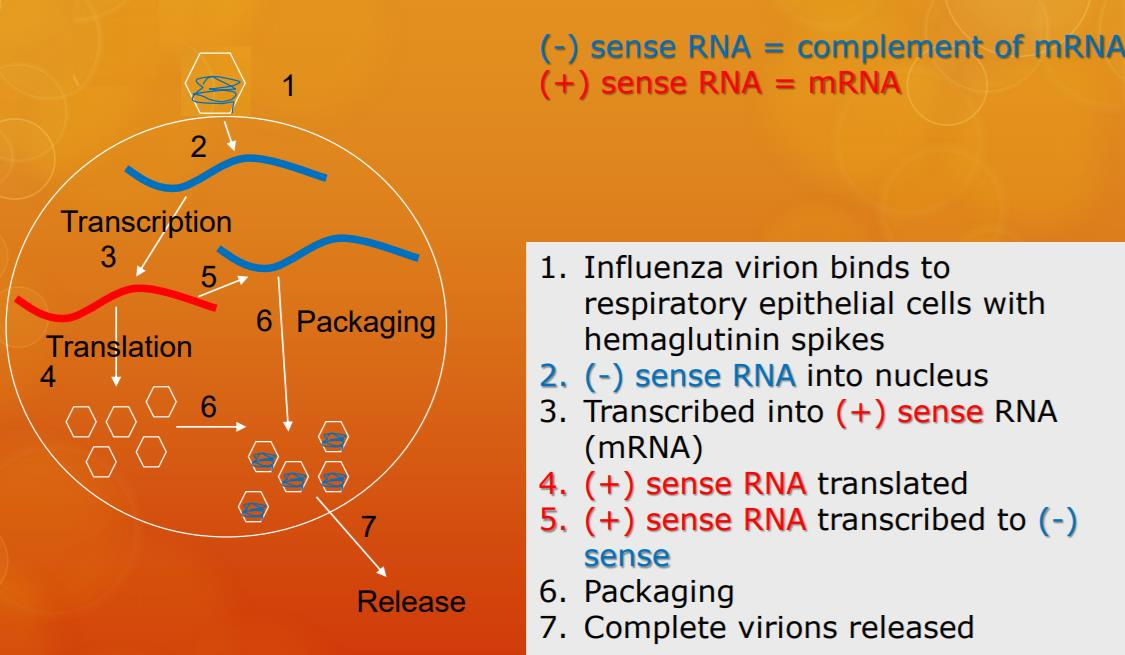
Lytic animal viruses follow similar infection stages to bacteriophages: attachment, penetration, biosynthesis, maturation, and release (see Figure \(\PageIndex{4}\)). However, the mechanisms of penetration, nucleic-acid biosynthesis, and release differ between bacterial and animal viruses. After binding to host receptors, animal viruses enter through endocytosis(engulfment by the host cell) or through membrane fusion (viral envelope with the host cell membrane). Many viruses are host specific, meaning they only infect a certain type of host; and most viruses only infect certain types of cells within tissues. This specificity is called a tissue tropism. Examples of this are demonstrated by the poliovirus, which exhibits tropism for the tissues of the brain and spinal cord, or the influenza virus, which has a primary tropism for the respiratory tract. The specificity of influenzavirus for the respiratory tract is because its glycoprotein hemagglutinin (HA) binds to receptors common on cells in the respiratory tract. Another protein on the surface of influenza virus, neuraminidase (NA), helps the virus access the cell surfaces by degrading the protective mucus.
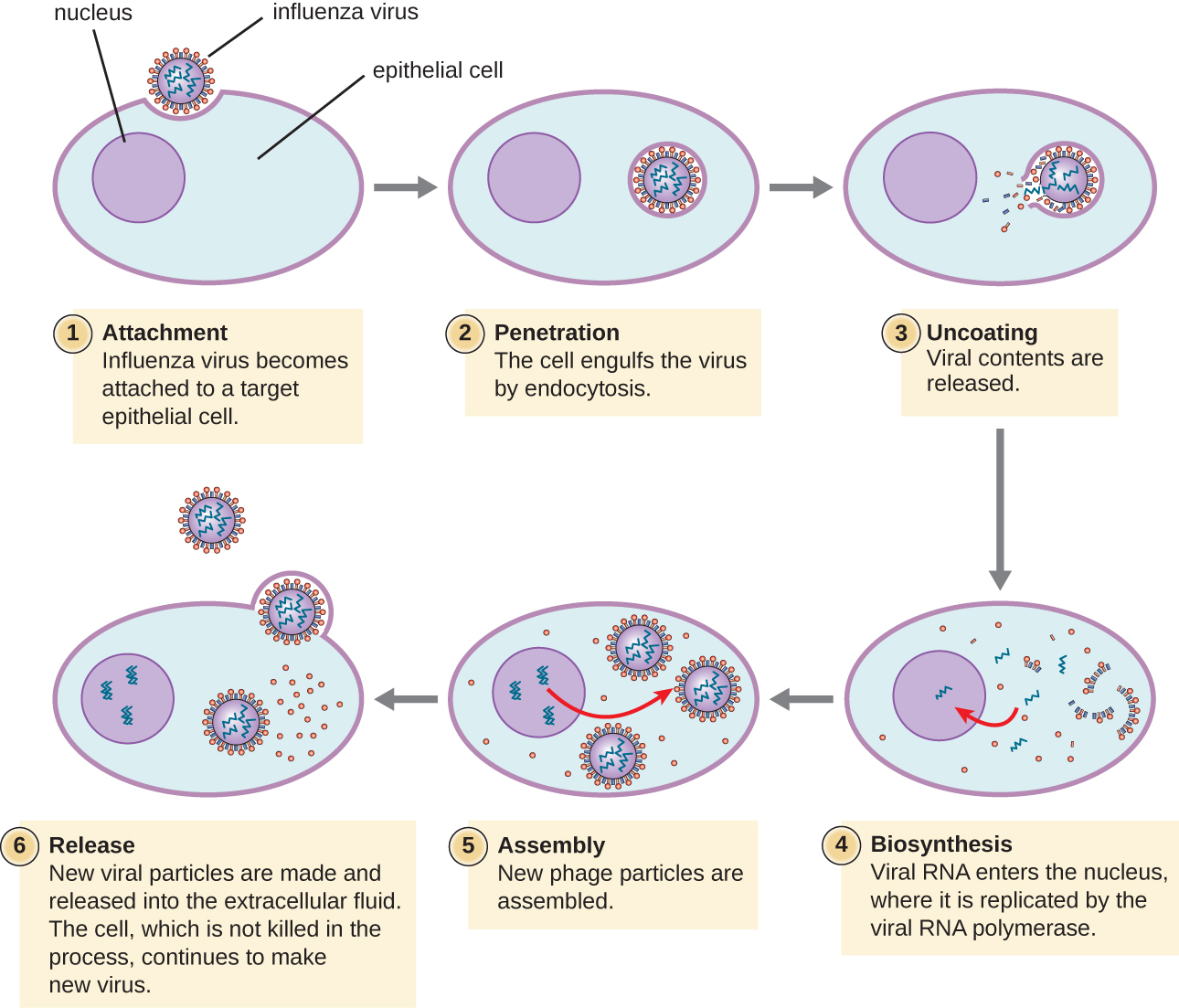
Animal viruses do not always express their genes using the normal flow of genetic information—from DNA to RNA to protein. Some viruses have a dsDNA genome like cellular organisms and can follow the normal flow. However, others may have ssDNA, dsRNA, or ssRNA genomes. The nature of the genome determines how the genome is replicated and expressed as viral proteins. If a genome is ssDNA, host enzymes will be used to synthesize a second strand that is complementary to the genome strand, thus producing dsDNA. The dsDNA can now be replicated, transcribed, and translated similar to host DNA.
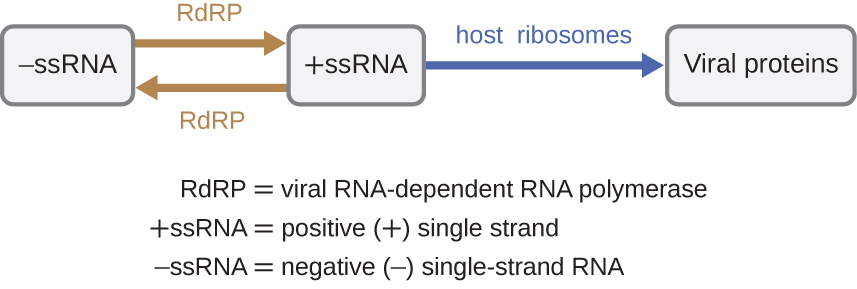
If the viral genome is RNA, a different mechanism must be used. There are three types of RNA genome: dsRNA, positive (+) single-strand (+ssRNA) or negative (−) single-strand RNA (−ssRNA). If a virus has a +ssRNA genome, it can be translated directly to make viral proteins. Viral genomic +ssRNA acts like cellular mRNA. However, if a virus contains a −ssRNA genome, the host ribosomes cannot translate it until the −ssRNA is replicated into +ssRNA by viral RNA-dependent RNA polymerase (RdRP) (see Figure \(\PageIndex{5}\)). The RdRP is brought in by the virus and can be used to make +ssRNA from the original −ssRNA genome. The RdRP is also an important enzyme for the replication of dsRNA viruses, because it uses the negative strand of the double-stranded genome as a template to create +ssRNA. The newly synthesized +ssRNA copies can then be translated by cellular ribosomes.
Retroviruses
An alternative mechanism for viral nucleic acid synthesis is observed in the retroviruses, which are +ssRNA viruses (see Figure \(\PageIndex{8}\)). Single-stranded RNA viruses such as HIV carry a special enzyme called reverse transcriptase within the capsid that synthesizes a complementary ssDNA (cDNA) copy using the +ssRNA genome as a template. The ssDNA is then made into dsDNA, which can integrate into the host chromosome and become a permanent part of the host. The integrated viral genome is called a provirus. The virus now can remain in the host for a long time to establish a chronic infection. The provirus stage is similar to the prophage stage in a bacterial infection during the lysogenic cycle. However, unlike prophage, the provirus does not undergo excision after splicing into the genome.

Exercise \(\PageIndex{3}\)
Is RNA-dependent RNA polymerase made from a viral gene or a host gene?
Persistent Infections
Persistent infection occurs when a virus is not completely cleared from the system of the host but stays in certain tissues or organs of the infected person. The virus may remain silent or undergo productive infection without seriously harming or killing the host. Mechanisms of persistent infection may involve the regulation of the viral or host gene expressions or the alteration of the host immune response. The two primary categories of persistent infections are latent infection and chronic infection. Examples of viruses that cause latent infections include herpes simplex virus (oral and genital herpes), varicella-zoster virus (chickenpox and shingles), and Epstein-Barr virus (mononucleosis). Hepatitis C virus and HIV are two examples of viruses that cause long-term chronic infections.
Latent Infection
Not all animal viruses undergo replication by the lytic cycle. There are viruses that are capable of remaining hidden or dormant inside the cell in a process called latency. These types of viruses are known as latent viruses and may cause latent infections. Viruses capable of latency may initially cause an acute infection before becoming dormant.
For example, the varicella-zoster virus infects many cells throughout the body and causes chickenpox, characterized by a rash of blisters covering the skin. About 10 to 12 days postinfection, the disease resolves and the virus goes dormant, living within nerve-cell ganglia for years. During this time, the virus does not kill the nerve cells or continue replicating. It is not clear why the virus stops replicating within the nerve cells and expresses few viral proteins but, in some cases, typically after many years of dormancy, the virus is reactivated and causes a new disease called shingles (Figure \(\PageIndex{9}\)). Whereas chickenpox affects many areas throughout the body, shingles is a nerve cell-specific disease emerging from the ganglia in which the virus was dormant.
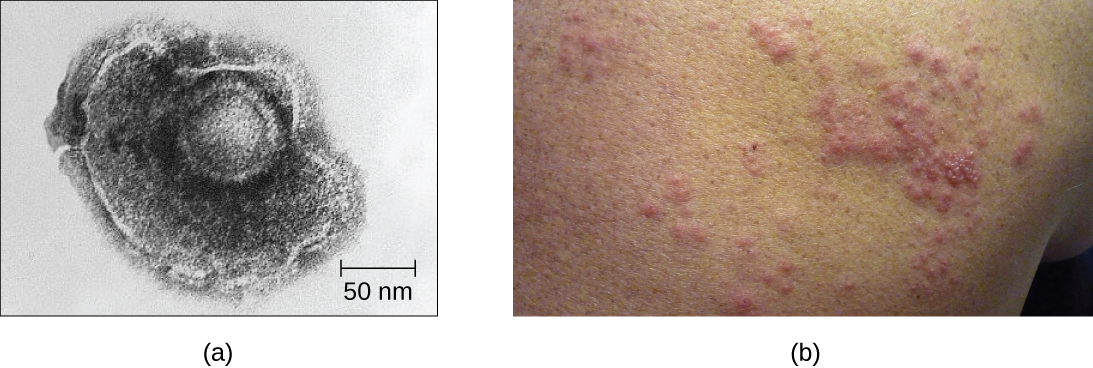
Latent viruses may remain dormant by existing as circular viral genome molecules outside of the host chromosome. Others become proviruses by integrating into the host genome. During dormancy, viruses do not cause any symptoms of disease and may be difficult to detect. A patient may be unaware that he or she is carrying the virus unless a viral diagnostic test has been performed.
Chronic Infection
A chronic infection is a disease with symptoms that are recurrent or persistent over a long time. Some viral infections can be chronic if the body is unable to eliminate the virus. HIV is an example of a virus that produces a chronic infection, often after a long period of latency. Once a person becomes infected with HIV, the virus can be detected in tissues continuously thereafter, but untreated patients often experience no symptoms for years. However, the virus maintains chronic persistence through several mechanisms that interfere with immune function, including preventing expression of viral antigens on the surface of infected cells, altering immune cells themselves, restricting expression of viral genes, and rapidly changing viral antigens through mutation. Eventually, the damage to the immune system results in progression of the disease leading to acquired immunodeficiency syndrome (AIDS). The various mechanisms that HIV uses to avoid being cleared by the immune system are also used by other chronically infecting viruses, including the hepatitis C virus.
Exercise \(\PageIndex{4}\)
In what two ways can a virus manage to maintain a persistent infection?
Summary
- Many viruses target specific hosts or tissues. Some may have more than one host.
- Many viruses follow several stages to infect host cells. These stages include attachment, penetration, uncoating, biosynthesis, maturation, and release.
- Bacteriophages have a lytic or lysogenic cycle. The lytic cycle leads to the death of the host, whereas the lysogenic cycle leads to integration of phage into the host genome.
- Bacteriophages inject DNA into the host cell, whereas animal viruses enter by endocytosis or membrane fusion.
- Animal viruses can undergo latency, similar to lysogeny for a bacteriophage.
- The majority of plant viruses are positive-strand ssRNA and can undergo latency, chronic, or lytic infection, as observed for animal viruses.
- The growth curve of bacteriophage populations is a one-step multiplication curve and not a sigmoidal curve, as compared to the bacterial growth curve.
- Bacteriophages transfer genetic information between hosts using either generalized or specialized transduction.
Footnotes
- 1 World Health Organization. “WHO Ebola Data and Statistics.” March 18, 2005. http://apps.who.int/gho/data/view.eb...150318?lang=en
Contributor
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)


