7.3: Alternate Forms of Catabolism: Fermentation, Lipids and Proteins
- Page ID
- 31812
Learning Objectives
- Define fermentation and explain why it does not require oxygen
- Describe the fermentation pathways and their end products and give examples of microorganisms that use these pathways
- Compare and contrast fermentation and anaerobic respiration
- Describe how lipids are catabolized
- Describe how lipid catabolism can be used to identify microbes
- Describe how proteins are catabolized
- Describe how protein catabolism can be used to identify bacteria
Fermentation
Many cells are unable to carry out respiration because of one or more of the following circumstances:
- The cell lacks a sufficient amount of any appropriate, inorganic, final electron acceptor to carry out cellular respiration.
- The cell lacks genes to make appropriate complexes and electron carriers in the electron transport system.
- The cell lacks genes to make one or more enzymes in the Krebs cycle.
Whereas lack of an appropriate inorganic final electron acceptor is environmentally dependent, the other two conditions are genetically determined. Thus, many prokaryotes, including members of the clinically important genus Streptococcus, are permanently incapable of respiration, even in the presence of oxygen. Conversely, many prokaryotes are facultative, meaning that, should the environmental conditions change to provide an appropriate inorganic final electron acceptor for respiration, organisms containing all the genes required to do so will switch to cellular respiration for glucose metabolism because respiration allows for much greater ATP production per glucose molecule.
If respiration does not occur, NADH must be reoxidized to NAD+ for reuse as an electron carrier for glycolysis, the cell’s only mechanism for producing any ATP, to continue. Some living systems use an organic molecule (commonly pyruvate) as a final electron acceptor through a process called fermentation. Fermentation does not involve an electron transport system and does not directly produce any additional ATP beyond that produced during glycolysis by substrate-level phosphorylation. Organisms carrying out fermentation, called fermenters, produce a maximum of two ATP molecules per glucose during glycolysis. Table \(\PageIndex{1}\) compares the final electron acceptors and methods of ATP synthesis in aerobic respiration, anaerobic respiration, and fermentation. Note that the number of ATP molecules shown for glycolysis assumes the Embden-Meyerhof-Parnas pathway. The number of ATP molecules made by substrate-level phosphorylation (SLP) versus oxidative phosphorylation (OP) are indicated.
| Type of Metabolism | Example | Final Electron Acceptor | Pathways Involved in ATP Synthesis (Type of Phosphorylation) | Maximum Yield of ATP Molecules |
|---|---|---|---|---|
| Aerobic respiration | Pseudomonas aeruginosa | \(\ce{O2}\) |
EMP glycolysis (SLP) Krebs cycle (SLP) Electron transport and chemiosmosis (OP): |
2 2 34 |
| Total | 38 | |||
| Anaerobic respiration | Paracoccus denitrificans |
\(\ce{NO3^{−}}\), \(\ce{SO^{−2}4}\), \(\ce{Fe^{+3}}\), \(\ce{CO2}\) other inorganics |
EMP glycolysis (SLP) Krebs cycle (SLP) Electron transport and chemiosmosis (OP): |
2 2 1–32 |
| Total | 5–36 | |||
| Fermentation | Candida albicans |
Organics (usually pyruvate) |
EMP glycolysis (SLP) Fermentation |
2 0 |
| Total | 2 |
Microbial fermentation processes have been manipulated by humans and are used extensively in the production of various foods and other commercial products, including pharmaceuticals. Microbial fermentation can also be useful for identifying microbes for diagnostic purposes.
Lactic Acid Fermentation
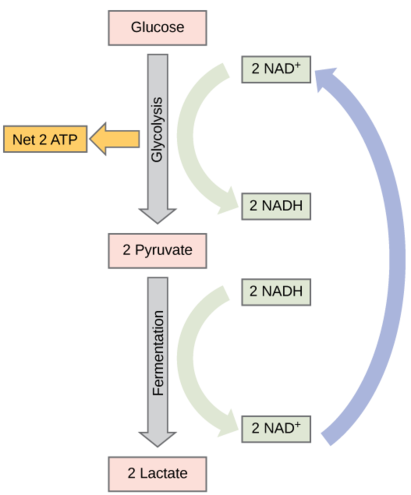
Fermentation by some bacteria, like those in yogurt and other soured food products, and by animals in muscles during oxygen depletion, is lactic acid fermentation. (Figure \(\PageIndex{1}\)). The chemical reaction of lactic acid fermentation is as follows:
Pyruvate + NADH↔lactic acid + NAD+Pyruvate + NADH ↔ lactic acid + NAD+
This type of fermentation is used routinely in mammalian red blood cells and in skeletal muscle that has an insufficient oxygen supply to allow aerobic respiration to continue (that is, in muscles used to the point of fatigue). In muscles, lactic acid accumulation must be removed by the blood circulation and the lactate brought to the liver for further metabolism.The enzyme used in this reaction is often lactate dehydrogenase (LDH). The reaction can proceed in either direction, but the reaction is inhibited by acidic conditions. Such lactic acid accumulation was once believed to cause muscle stiffness, fatigue, and soreness, although more recent research disputes this hypothesis. Once the lactic acid has been removed from the muscle and circulated to the liver, it can be reconverted into pyruvic acid and further catabolized for energy.
Bacteria of several gram-positive genera, including Lactobacillus, Leuconostoc, and Streptococcus, are collectively known as the lactic acid bacteria (LAB), and various strains are important in food production. During yogurt and cheese production, the highly acidic environment generated by lactic acid fermentation denatures proteins contained in milk, causing it to solidify. When lactic acid is the only fermentation product, the process is said to be homolactic fermentation; such is the case for Lactobacillus delbrueckii and S. thermophiles used in yogurt production. However, many bacteria perform heterolactic fermentation, producing a mixture of lactic acid, ethanol and/or acetic acid, and CO2 as a result, because of their use of the branched pentose phosphate pathway instead of the EMP pathway for glycolysis. One important heterolactic fermenter is Leuconostoc mesenteroides, which is used for souring vegetables like cucumbers and cabbage, producing pickles and sauerkraut, respectively.
Lactic acid bacteria are also important medically. The production of low pH environments within the body inhibits the establishment and growth of pathogens in these areas. For example, the vaginal microbiota is composed largely of lactic acid bacteria, but when these bacteria are reduced, yeast can proliferate, causing a yeast infection. Additionally, lactic acid bacteria are important in maintaining the health of the gastrointestinal tract and, as such, are the primary component of probiotics.
Alcohol Fermentation
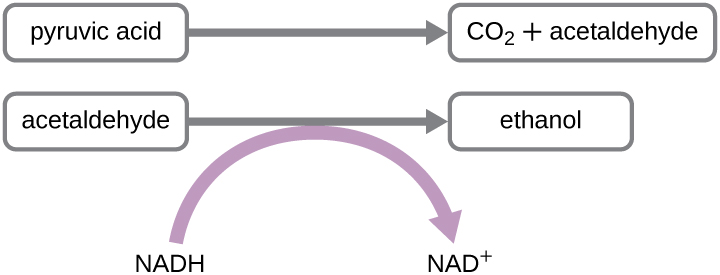
Another familiar fermentation process is alcohol fermentation, which produces ethanol. The ethanol fermentation reaction is shown in Figure \(\PageIndex{2}\). In the first reaction, the enzyme pyruvate decarboxylase removes a carboxyl group from pyruvate, releasing CO2 gas while producing the two-carbon molecule acetaldehyde. The second reaction, catalyzed by the enzyme alcohol dehydrogenase, transfers an electron from NADH to acetaldehyde, producing ethanol and NAD+. The ethanol fermentation of pyruvate by the yeast Saccharomyces cerevisiae is used in the production of alcoholic beverages and also makes bread products rise due to CO2 production. Ethanol tolerance of yeast is variable, ranging from about 5 percent to 21 percent, depending on the yeast strain and environmental conditions. Outside of the food industry, ethanol fermentation of plant products is important in biofuel production.
Other Fermentation Pathways
Beyond lactic acid fermentation and alcohol fermentation, many other fermentation methods occur in prokaryotes, all for the purpose of ensuring an adequate supply of NAD+ for glycolysis (Table \(\PageIndex{2}\)). Without these pathways, glycolysis would not occur and no ATP would be harvested from the breakdown of glucose. It should be noted that most forms of fermentation besides homolactic fermentation produce gas, commonly CO2 and/or hydrogen gas. Many of these different types of fermentation pathways are also used in food production and each results in the production of different organic acids, contributing to the unique flavor of a particular fermented food product. The propionic acid produced during propionic acid fermentation contributes to the distinctive flavor of Swiss cheese, for example.
Several fermentation products are important commercially outside of the food industry. For example, chemical solvents such as acetone and butanol are produced during acetone-butanol-ethanol fermentation. Complex organic pharmaceutical compounds used in antibiotics (e.g., penicillin), vaccines, and vitamins are produced through mixed acid fermentation. Fermentation products are used in the laboratory to differentiate various bacteria for diagnostic purposes. For example, enteric bacteria are known for their ability to perform mixed acid fermentation, reducing the pH, which can be detected using a pH indicator. Similarly, the bacterial production of acetoin during butanediol fermentation can also be detected. Gas production from fermentation can also be seen in an inverted Durham tube that traps produced gas in a broth culture.
Microbes can also be differentiated according to the substrates they can ferment. For example, E. coli can ferment lactose, forming gas, whereas some of its close gram-negative relatives cannot. The ability to ferment the sugar alcohol sorbitol is used to identify the pathogenic enterohemorrhagic O157:H7 strain of E. coli because, unlike other E. coli strains, it is unable to ferment sorbitol. Last, mannitol fermentation differentiates the mannitol-fermenting Staphylococcus aureus from other non–mannitol-fermenting staphylococci.
| Pathway | End Products | Example Microbes | Commercial Products |
|---|---|---|---|
| Acetone-butanol-ethanol | Acetone, butanol, ethanol, CO2 | Clostridium acetobutylicum | Commercial solvents, gasoline alternative |
| Alcohol | Ethanol, CO2 | Candida, Saccharomyces | Beer, bread |
| Butanediol | Formic and lactic acid; ethanol; acetoin; 2,3 butanediol; CO2; hydrogen gas | Klebsiella, Enterobacter | Chardonnay wine |
| Butyric acid | Butyric acid, CO2, hydrogen gas | Clostridium butyricum | Butter |
| Lactic acid | Lactic acid | Streptococcus, Lactobacillus | Sauerkraut, yogurt, cheese |
| Mixed acid | Acetic, formic, lactic, and succinic acids; ethanol, CO2, hydrogen gas | Escherichia, Shigella | Vinegar, cosmetics, pharmaceuticals |
| Propionic acid | Acetic acid, propionic acid, CO2 | Propionibacterium, Bifidobacterium | Swiss cheese |
Exercise \(\PageIndex{5}\)
When would a metabolically versatile microbe perform fermentation rather than cellular respiration?
No Carbohydrate Available
Previous sections have discussed the catabolism of glucose, which provides energy to living cells, as well as how polysaccharides like glycogen, starch, and cellulose are degraded to glucose monomers. But microbes consume more than just carbohydrates for food. In fact, the microbial world is known for its ability to degrade a wide range of molecules, both naturally occurring and those made by human processes, for use as carbon sources. In this section, we will see that the pathways for both lipid and protein catabolism connect to those used for carbohydrate catabolism, eventually leading into glycolysis, the transition reaction, and the Krebs cycle pathways. Metabolic pathways should be considered to be porous—that is, substances enter from other pathways, and intermediates leave for other pathways. These pathways are not closed systems. Many of the substrates, intermediates, and products in a particular pathway are reactants in other pathways.
Lipid Catabolism
Triglycerides are a form of long-term energy storage in animals. They are made of glycerol and three fatty acids. Phospholipids compose the cell and organelle membranes of all organisms except the archaea. Phospholipid structure is similar to triglycerides except that one of the fatty acids is replaced by a phosphorylated head group. Triglycerides and phospholipids are broken down first by releasing fatty acid chains (and/or the phosphorylated head group, in the case of phospholipids) from the three-carbon glycerol backbone. The reactions breaking down triglycerides are catalyzed by lipases and those involving phospholipids are catalyzed by phospholipases. These enzymes contribute to the virulence of certain microbes, such as the bacterium Staphylococcus aureus and the fungus Cryptococcus neoformans. These microbes use phospholipases to destroy lipids and phospholipids in host cells and then use the catabolic products for energy.
The resulting products of lipid catabolism, glycerol and fatty acids, can be further degraded. Glycerol can be phosphorylated to glycerol-3-phosphate and easily converted to glyceraldehyde 3-phosphate, which continues through glycolysis. The released fatty acids are catabolized in a process called β-oxidation, which sequentially removes two-carbon acetyl groups from the ends of fatty acid chains, reducing NAD+ and FAD to produce NADH and FADH2, respectively, whose electrons can be used to make ATP by oxidative phosphorylation. The acetyl groups produced during β-oxidation are carried by coenzyme A to the Krebs cycle, and their movement through this cycle results in their degradation to CO2, producing ATP by substrate-level phosphorylation and additional NADH and FADH2 molecules.
Other types of lipids can also be degraded by certain microbes. For example, the ability of certain pathogens, like Mycobacterium tuberculosis, to degrade cholesterol contributes to their virulence. The side chains of cholesterol can be easily removed enzymatically, but degradation of the remaining fused rings is more problematic. The four fused rings are sequentially broken in a multistep process facilitated by specific enzymes, and the resulting products, including pyruvate, can be further catabolized in the Krebs cycle.
Exercise \(\PageIndex{1}\)
How can lipases and phospholipases contribute to virulence in microbes?
Protein Catabolism
Proteins are degraded through the concerted action of a variety of microbial protease enzymes. Extracellular proteases cut proteins internally at specific amino acid sequences, breaking them down into smaller peptides that can then be taken up by cells. Some clinically important pathogens can be identified by their ability to produce a specific type of extracellular protease. For example, the production of the extracellular protease gelatinase by members of the genera Proteus and Serratia can be used to distinguish them from other gram-negative enteric bacteria. Following inoculation and growth of microbes in gelatin broth, degradation of the gelatin protein due to gelatinase production prevents solidification of gelatin when refrigerated. Other pathogens can be distinguished by their ability to degrade casein, the main protein found in milk. When grown on skim milk agar, production of the extracellular protease caseinase causes degradation of casein, which appears as a zone of clearing around the microbial growth. Caseinase production by the opportunist pathogen Pseudomonas aeruginosa can be used to distinguish it from other related gram-negative bacteria.
After extracellular protease degradation and uptake of peptides in the cell, the peptides can then be broken down further into individual amino acids by additional intracellular proteases, and each amino acid can be enzymatically deaminated to remove the amino group. The remaining molecules can then enter the transition reaction or the Krebs cycle.
Exercise \(\PageIndex{2}\)
How can protein catabolism help identify microbes?
Clinical Focus: part 3
Because bacterial meningitis progresses so rapidly, Hannah’s doctors had decided to treat her aggressively with antibiotics, based on empirical observation of her symptoms. However, laboratory testing to confirm the cause of Hannah’s meningitis was still important for several reasons. N. meningitidis is an infectious pathogen that can be spread from person to person through close contact; therefore, if tests confirm N. meningitidis as the cause of Hannah’s symptoms, Hannah’s parents and others who came into close contact with her might need to be vaccinated or receive prophylactic antibiotics to lower their risk of contracting the disease. On the other hand, if it turns out that N. meningitidis is not the cause, Hannah’s doctors might need to change her treatment.
The clinical laboratory performed a Gram stain on Hannah’s blood and CSF samples. The Gram stain showed the presence of a bean-shaped gram-negative diplococcus. The technician in the hospital lab cultured Hannah’s blood sample on both blood agar and chocolate agar, and the bacterium that grew on both media formed gray, nonhemolytic colonies. Next, he performed an oxidase test on this bacterium and determined that it was oxidase positive. Last, he examined the repertoire of sugars that the bacterium could use as a carbon source and found that the bacterium was positive for glucose and maltose use but negative for lactose and sucrose use. All of these test results are consistent with characteristics of N. meningitidis.
Exercise \(\PageIndex{3}\)
- What do these test results tell us about the metabolic pathways of N. meningitidis?
- Why do you think that the hospital used these biochemical tests for identification in lieu of molecular analysis by DNA testing?
Key Concepts and Summary
- Fermentation uses an organic molecule as a final electron acceptor to regenerate NAD+ from NADH so that glycolysis can continue.
- Fermentation does not involve an electron transport system, and no ATP is made by the fermentation process directly. Fermenters make very little ATP—only two ATP molecules per glucose molecule during glycolysis.
- Microbial fermentation processes have been used for the production of foods and pharmaceuticals, and for the identification of microbes.
- During lactic acid fermentation, pyruvate accepts electrons from NADH and is reduced to lactic acid. Microbes performing homolactic fermentation produce only lactic acid as the fermentation product; microbes performing heterolactic fermentation produce a mixture of lactic acid, ethanol and/or acetic acid, and CO2.
- Lactic acid production by the normal microbiota prevents growth of pathogens in certain body regions and is important for the health of the gastrointestinal tract.
- During ethanol fermentation, pyruvate is first decarboxylated (releasing CO2) to acetaldehyde, which then accepts electrons from NADH, reducing acetaldehyde to ethanol. Ethanol fermentation is used for the production of alcoholic beverages, for making bread products rise, and for biofuel production.
- Fermentation products of pathways (e.g., propionic acid fermentation) provide distinctive flavors to food products. Fermentation is used to produce chemical solvents (acetone-butanol-ethanol fermentation) and pharmaceuticals (mixed acid fermentation).
- Specific types of microbes may be distinguished by their fermentation pathways and products. Microbes may also be differentiated according to the substrates they are able to ferment.
- Collectively, microbes have the ability to degrade a wide variety of carbon sources besides carbohydrates, including lipids and proteins. The catabolic pathways for all of these molecules eventually connect into glycolysis and the Krebs cycle.
- Several types of lipids can be microbially degraded. Triglycerides are degraded by extracellular lipases, releasing fatty acids from the glycerol backbone. Phospholipids are degraded by phospholipases, releasing fatty acids and the phosphorylated head group from the glycerol backbone. Lipases and phospholipases act as virulence factors for certain pathogenic microbes.
- Fatty acids can be further degraded inside the cell through β-oxidation, which sequentially removes two-carbon acetyl groups from the ends of fatty acid chains.
- Protein degradation involves extracellular proteases that degrade large proteins into smaller peptides. Detection of the extracellular proteases gelatinase and caseinase can be used to differentiate clinically relevant bacteria.
Contributors and Attributions
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)


