4.2: Classifying Prokaryotes and Examples
- Page ID
- 31781
Learning Objectives
- Explain how prokaryotes are classified what criteria are used to help group them into phylum
- Highlight some interesting and diverse prokaryotes
Taxonomy and Systematics
Assigning prokaryotes to a certain species is challenging. They do not reproduce sexually, so it is not possible to classify them according to the presence or absence of interbreeding. Also, they do not have many morphological features. Traditionally, the classification of prokaryotes was based on their shape, staining patterns, and biochemical or physiological differences. More recently, as technology has improved, the nucleotide sequences in genes (particularly rRNA) have become an important criterion of microbial classification.
In 1923, American microbiologist David Hendricks Bergey (1860–1937) published A Manual in Determinative Bacteriology. With this manual, he attempted to summarize the information about the kinds of bacteria known at that time, using Latin binomial classification. Bergey also included the morphological, physiological, and biochemical properties of these organisms. His manual has been updated multiple times to include newer bacteria and their properties. It is a great aid in bacterial taxonomy and methods of characterization of bacteria. A more recent sister publication, the five-volume Bergey’s Manual of Systematic Bacteriology, expands on Bergey’s original manual. It includes a large number of additional species, along with up-to-date descriptions of the taxonomy and biological properties of all named prokaryotic taxa. This publication incorporates the approved names of bacteria as determined by the List of Prokaryotic Names with Standing in Nomenclature (LPSN). It can also be found as a living document, available through subscription, to try and keep it as up-to-date as possible.
Classification by Staining Patterns
According to their staining patterns, which depend on the properties of their cell walls, bacteria have traditionally been classified into gram-positive, gram-negative, and “atypical,” meaning neither gram-positive nor gram-negative. As explained in earlier, gram-positive bacteria possess a thick peptidoglycan cell wall that retains the primary stain (crystal violet) during the decolorizing step; they remain purple after the gram-stain procedure because the crystal violet dominates the light red/pink color of the secondary counterstain, safranin. In contrast, gram-negative bacteria possess a thin peptidoglycan cell wall that does not prevent the crystal violet from washing away during the decolorizing step; therefore, they appear light red/pink after staining with the safranin. Bacteria that cannot be stained by the standard Gram stain procedure are called atypical bacteria. Included in the atypical category are species of Mycoplasma and Chlamydia, which lack a cell wall and therefore cannot retain the gram-stain reagents. Rickettsia are also considered atypical because they are too small to be evaluated by the Gram stain.
More recently, scientists have begun to further classify gram-negative and gram-positive bacteria. They have added a special group of deeply branching bacteria based on a combination of physiological, biochemical, and genetic features. They also now further classify gram-negative bacteria into Proteobacteria, Cytophaga-Flavobacterium-Bacteroides (CFB), and spirochetes.
The deeply branching bacteria are thought to be a very early evolutionary form of bacteria. They live in hot, acidic, ultraviolet-light-exposed, and anaerobic (deprived of oxygen) conditions. Proteobacteria is a phylum of very diverse groups of gram-negative bacteria; it includes some important human pathogens (e.g., E. coli and Bordetella pertussis). The CFB group of bacteria includes components of the normal human gut microbiota, like Bacteroides. The spirochetes are spiral-shaped bacteria and include the pathogen Treponema pallidum, which causes syphilis. We will characterize these groups of bacteria in more detail later in this section.
Based on their prevalence of guanine and cytosine nucleotides, gram-positive bacteria are also classified into low G+C and high G+C gram-positive bacteria. The low G+C gram-positive bacteria have less than 50% of guanine and cytosine nucleotides in their DNA. They include human pathogens, such as those that cause anthrax (Bacillus anthracis), tetanus (Clostridium tetani), and listeriosis (Listeria monocytogenes). High G+C gram-positive bacteria, which have more than 50% guanine and cytosine nucleotides in their DNA, include the bacteria that cause diphtheria (Corynebacterium diphtheriae), tuberculosis (Mycobacterium tuberculosis), and other diseases.
Using these two descriptions however, will only get small amounts of separation, so microbiologist turn to two other descriptions: the microbes interaction with or reliance on oxygen gas, and their main mode of gaining of nutrients (both more fully described in chapters 7 and 8). The classifications of prokaryotes are constantly changing as new species are being discovered and new information comes to light. Included in the next section are some highlights from the different major groups and phylum of the Bacterial and Archaeal domains.
Exercise \(\PageIndex{1}\)
What characteristics do scientists use to classify prokaryotes?
Proteobacteria
In 1987, the American microbiologist Carl Woese (1928–2012) suggested that a large and diverse group of bacteria that he called “purple bacteria and their relatives” should be defined as a separate phylum within the domain Bacteria based on the similarity of the nucleotide sequences in their genome.1 This phylum of gram-negative bacteria subsequently received the name Proteobacteria. It includes many bacteria that are part of the normal human microbiota as well as many pathogens. The Proteobacteria are further divided into five classes: Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, and Epsilonproteobacteria.
Alphaproteobacteria include intracellular parasites
Among the Alphaproteobacteria are two taxa, chlamydias and rickettsias, that are obligate intracellular pathogens, meaning that part of their life cycle must occur inside other cells called host cells. When not growing inside a host cell, Chlamydia and Rickettsia are metabolically inactive outside of the host cell. They cannot synthesize their own adenosine triphosphate (ATP), and, therefore, rely on cells for their energy needs.
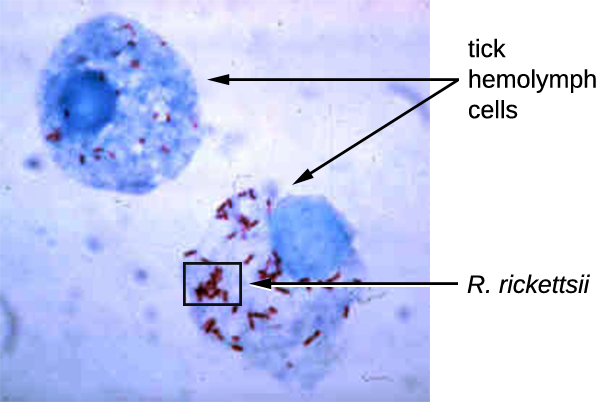
Rickettsia spp. include a number of serious human pathogens. For example, R. rickettsii causes Rocky Mountain spotted fever, a life-threatening form of meningoencephalitis (inflammation of the membranes that wrap the brain). R. rickettsii infects ticks and can be transmitted to humans via a bite from an infected tick (Figure \(\PageIndex{1}\)). Another species of Rickettsia, R. prowazekii, is spread by lice. It causes epidemic typhus, a severe infectious disease common during warfare and mass migrations of people. R. prowazekii infects human endothelium cells, causing inflammation of the inner lining of blood vessels, high fever, abdominal pain, and sometimes delirium. A relative, R. typhi, causes a less severe disease known as murine or endemic typhus, which is still observed in the southwestern United States during warm seasons.
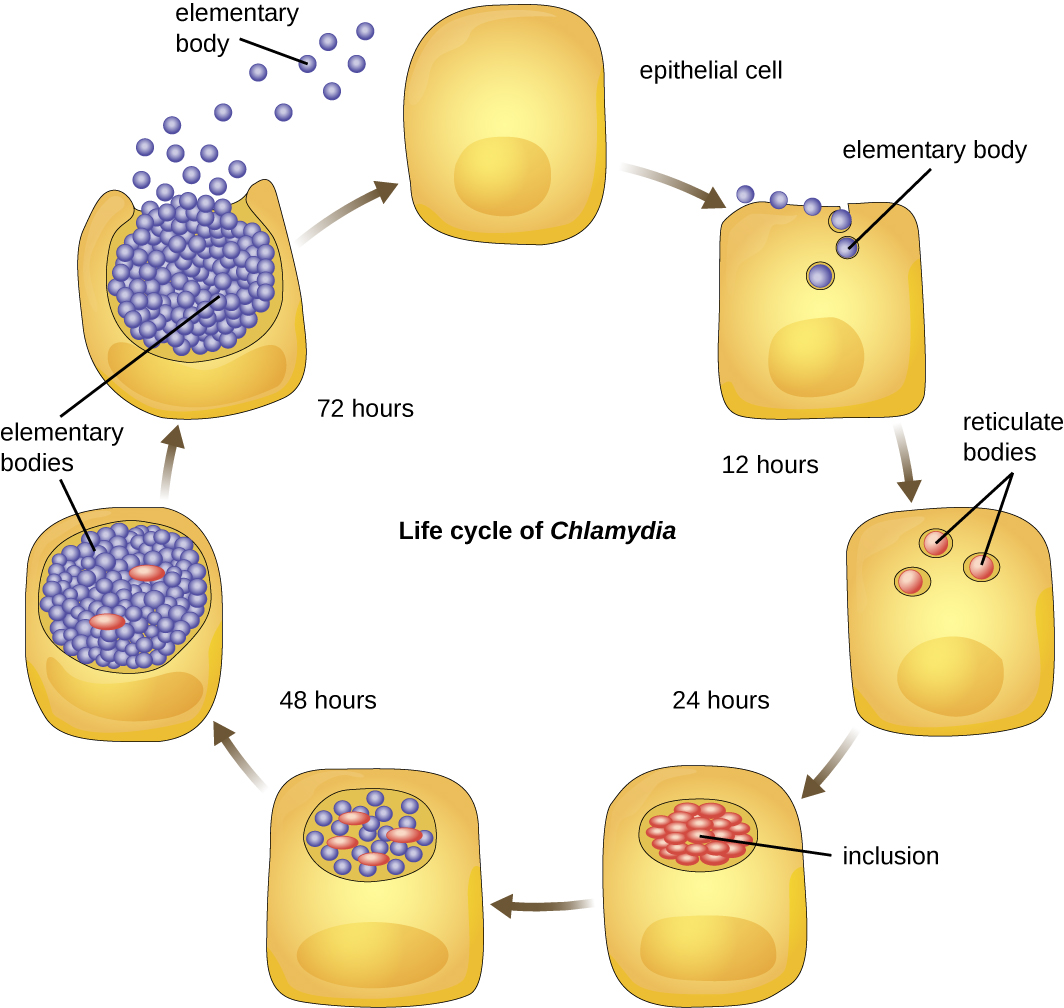
Chlamydia is another taxon of the Alphaproteobacteria. Members of this genus are extremely resistant to the cellular defenses, giving them the ability to spread from host to host rapidly via elementary bodies. The metabolically and reproductively inactive elementary bodies are the endospore-like form of intracellular bacteria that enter an epithelial cell, where they become active. Figure \(\PageIndex{2}\) illustrates the life cycle of Chlamydia. C. trachomatis is a human pathogen that causes trachoma, a disease of the eyes, often leading to blindness. C. trachomatis also causes the sexually transmitted disease lymphogranuloma venereum (LGV). This disease is often mildly symptomatic, manifesting as regional lymph node swelling, or it may be asymptomatic, but it is extremely contagious and is common on college campuses.
Betaproteobacteria include other familiar infectious bacteria
Unlike Alphaproteobacteria, which survive on a minimal amount of nutrients, the class Betaproteobacteria are eutrophs (or copiotrophs), meaning that they require a copious amount of organic nutrients. Betaproteobacteria often grow between aerobic and anaerobic areas (e.g., in mammalian intestines). Some genera include species that are human pathogens, able to cause severe, sometimes life-threatening disease. The pathogen responsible for pertussis (whooping cough) is also a member of Betaproteobacteria. The bacterium Bordetella pertussis, from the order Burkholderiales, produces several toxins that paralyze the movement of cilia in the human respiratory tract and directly damage cells of the respiratory tract, causing a severe cough.
The genus Neisseria, for example, includes the bacteria N. gonorrhoeae, the causative agent of the STI gonorrhea, and N. meningitides, the causative agent of bacterial meningitis. Neisseria are cocci that live on mucosal surfaces of the human body. They are fastidious, or difficult to culture, and they require high levels of moisture, nutrient supplements, and carbon dioxide. Also, Neisseria are microaerophilic, meaning that they require low levels of oxygen. For optimal growth and for the purposes of identification, Neisseria spp. are grown on chocolate agar (i.e., agar supplemented by partially hemolyzed red blood cells). Their characteristic pattern of growth in culture is diplococcal: pairs of cells resembling coffee beans (Figure \(\PageIndex{3}\)).
Gammaproteobacteria
The most diverse class of gram-negative bacteria is Gammaproteobacteria, and it includes a number of human pathogens. For example, a large and diverse family, Pseudomonaceae, includes the genus Pseudomonas. Within this genus is the species P. aeruginosa, a pathogen responsible for diverse infections in various regions of the body. It often infects wounds and burns, can be the cause of chronic urinary tract infections, and can be an important cause of respiratory infections in patients with cystic fibrosis or patients on mechanical ventilators. Infections by P. aeruginosa are often difficult to treat because the bacterium is resistant to many antibiotics and has a remarkable ability to form biofilms. Other representatives of Pseudomonas include the fluorescent (glowing) bacterium P. fluorescens and the soil bacteria P. putida, which is known for its ability to degrade xenobiotics (substances not naturally produced or found in living organisms).
Another famous member is E. coli has been perhaps the most studied bacterium since it was first described in 1886 by Theodor Escherich (1857–1911). Many strains of E. coli are in mutualistic relationships with humans. However, some strains produce a potentially deadly toxin called Shiga toxin, which perforates cellular membranes in the large intestine, causing bloody diarrhea and peritonitis (inflammation of the inner linings of the abdominal cavity). Other E. coli strains may cause traveler’s diarrhea, a less severe but very widespread disease. The genera Pasteruella, Legionella, Haemophilus and Salmonella.
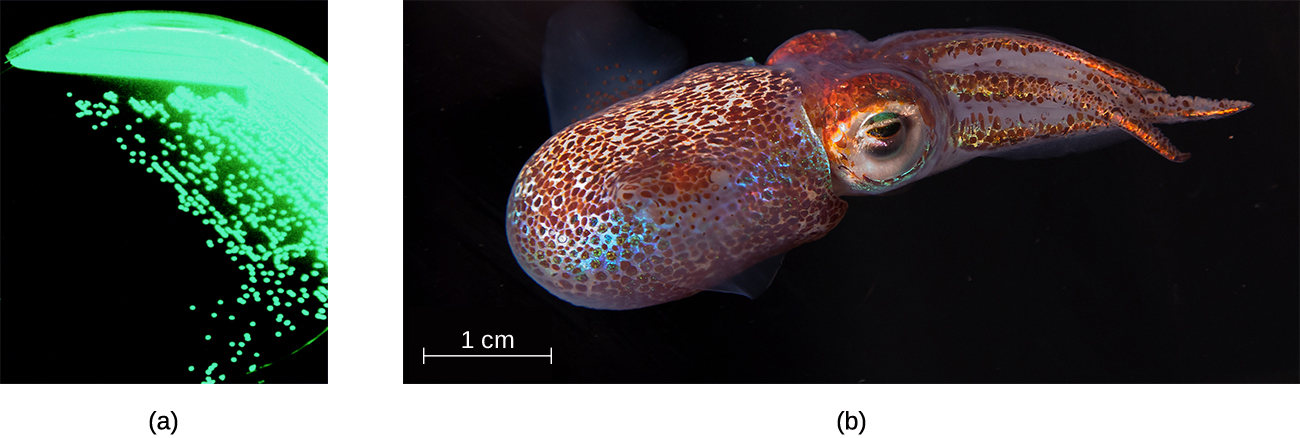
The order Vibrionales includes the human pathogen Vibrio cholerae. This comma-shaped aquatic bacterium thrives in highly alkaline environments like shallow lagoons and sea ports. A toxin produced by V. cholerae causes hypersecretion of electrolytes and water in the large intestine, leading to profuse watery diarrhea and dehydration. V. parahaemolyticus is also a cause of gastrointestinal disease in humans, whereas V. vulnificus causes serious and potentially life-threatening cellulitis (infection of the skin and deeper tissues) and blood-borne infections. Another representative of Vibrionales, Aliivibrio fischeri, engages in a symbiotic relationship with squid. The squid provides nutrients for the bacteria to grow and the bacteria produce bioluminescence that protects the squid from predators (Figure \(\PageIndex{4}\)).
Deltaproteobacteria include potentially helpful a scattering of helpful bacteria
The Deltaproteobacteria is a small class of gram-negative Proteobacteria that includes sulfate-reducing bacteria(SRBs), so named because they use sulfate as the final electron acceptor in the electron transport chain. Few SRBs are pathogenic. However, the SRB Desulfovibrio orale is associated with periodontal disease (disease of the gums).
Deltaproteobacteria also includes the genus Bdellovibrio, species of which are parasites of other gram-negative bacteria. Bdellovibrio invades the cells of the host bacterium, positioning itself in the periplasm, the space between the plasma membrane and the cell wall, feeding on the host’s proteins and polysaccharides. The infection is lethal for the host cells.
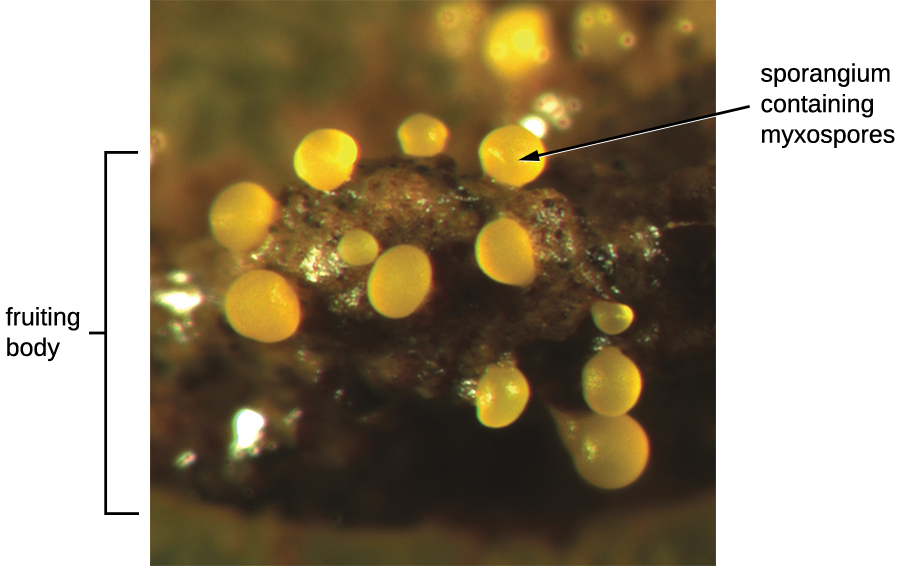
Another type of Deltaproteobacteria, myxobacteria, lives in the soil, scavenging inorganic compounds. Motile and highly social, they interact with other bacteria within and outside their own group. They can form multicellular, macroscopic “fruiting bodies” (Figure \(\PageIndex{5}\)), structures that are still being studied by biologists and bacterial ecologists.2 These bacteria can also form metabolically inactive myxospores.
Epsilonproteobacteria includes familiar GI pathogens
The smallest class of Proteobacteria is Epsilonproteobacteria, which are gram-negative microaerophilic bacteria (meaning they only require small amounts of oxygen in their environment). Two clinically relevant genera of Epsilonproteobacteria are Campylobacter and Helicobacter, both of which include human pathogens. Campylobacter can cause food poisoning that manifests as severe enteritis (inflammation in the small intestine). This condition, caused by the species C. jejuni, is rather common in developed countries, usually because of eating contaminated poultry products. Chickens often harbor C. jejuni in their gastrointestinal tract and feces, and their meat can become contaminated during processing.
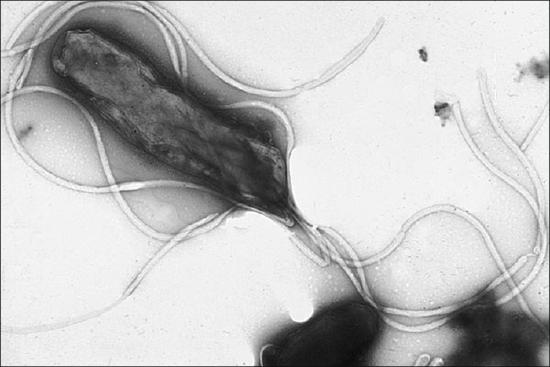
Within the genus Helicobacter, the helical, flagellated bacterium H. pylori has been identified as a beneficial member of the stomach microbiota, but it is also the most common cause of chronic gastritis and ulcers of the stomach and duodenum (Figure \(\PageIndex{6}\)). Studies have also shown that H. pylori is linked to stomach cancer.3 H. pylori is somewhat unusual in its ability to survive in the highly acidic environment of the stomach. It produces urease and other enzymes that modify its environment to make it less acidic.
Nonproteobacteria Gram-negative Bacteria and Phototrophic Bacteria
The majority of the gram-negative bacteria belong to the phylum Proteobacteria, discussed in the previous section. Those that do not are called the nonproteobacteria. In this section, we will describe three classes of gram-negative nonproteobacteria: the spirochetes, the CFB group, and the Planctomycetes. A diverse group of phototrophic bacteria that includes Proteobacteria and nonproteobacteria will be discussed at the end of this section.
Spirochetes
Several genera of spirochetes include human pathogens. For example, the genus Treponema includes a species T. pallidum, which is further classified into four subspecies: T. pallidum pallidum, T. pallidum pertenue, T. pallidum carateum, and T. pallidum endemicum. The subspecies T. pallidum pallidum causes the sexually transmitted infection known as syphilis, the third most prevalent sexually transmitted bacterial infection in the United States, after chlamydia and gonorrhea. The other subspecies of T. pallidum cause tropical infectious diseases of the skin, bones, and joints.
Another genus of spirochete, Borrelia, contains a number of pathogenic species. B. burgdorferi causes Lyme disease, which is transmitted by several genera of ticks (notably Ixodes and Amblyomma) and often produces a “bull’s eye” rash, fever, fatigue, and, sometimes, debilitating arthritis. B. recurrens causes a condition known as relapsing fever.
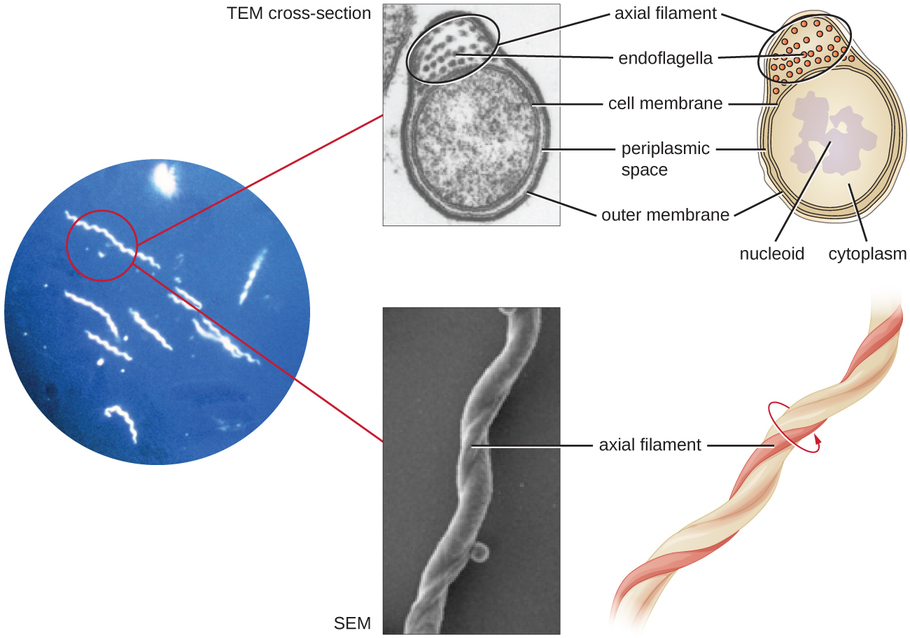
Spirochetes are characterized by their long (up to 250 μm), spiral-shaped bodies. Most spirochetes are also very thin, which makes it difficult to examine gram-stained preparations under a conventional brightfield microscope. Darkfield fluorescent microscopy is typically used instead. Spirochetes are also difficult or even impossible to culture. They are highly motile, using their axial filament to propel themselves. The axial filament is similar to a flagellum, but it wraps around the cell and runs inside the cell body of a spirochete in the periplasmic space between the outer membrane and the plasma membrane (Figure \(\PageIndex{7}\)).
Phototrophic Bacteria
The phototrophic bacteria are a large and diverse category of bacteria that do not represent a taxon but, rather, a group of bacteria that use sunlight as their primary source of energy. This group contains both Proteobacteria and nonproteobacteria. They use solar energy to synthesize ATP through photosynthesis. When they produce oxygen, they perform oxygenic photosynthesis. When they do not produce oxygen, they perform anoxygenic photosynthesis. With the exception of some cyanobacteria, the majority of phototrophic bacteria perform anoxygenic photosynthesis.
One large group of phototrophic bacteria includes the purple or green bacteria that perform photosynthesis with the help of bacteriochlorophylls, which are green, purple, or blue pigments similar to chlorophyll in plants. Some of these bacteria have a varying amount of red or orange pigments called carotenoids. Their color varies from orange to red to purple to green (Figure \(\PageIndex{8}\)), and they are able to absorb light of various wavelengths. Traditionally, these bacteria are classified into sulfur and nonsulfur bacteria; they are further differentiated by color. The sulfur bacteria perform anoxygenic photosynthesis, using sulfites as electron donors and releasing free elemental sulfur. Nonsulfur bacteria use organic substrates, such as succinate and malate, as donors of electrons.

Another large, diverse group of phototrophic bacteria compose the phylum Cyanobacteria; they get their blue-green color from the chlorophyll contained in their cells (Figure \(\PageIndex{9}\)). Species of this group perform oxygenic photosynthesis, producing megatons of gaseous oxygen. Scientists hypothesize that cyanobacteria played a critical role in the change of our planet’s anoxic atmosphere 1–2 billion years ago to the oxygen-rich environment we have today.4 Cyanobacteria have other remarkable properties. Amazingly adaptable, they thrive in many habitats, including marine and freshwater environments, soil, and even rocks. They can live at a wide range of temperatures, even in the extreme temperatures of the Antarctic. They can live as unicellular organisms or in colonies, and they can be filamentous, forming sheaths or biofilms. Many of them fix nitrogen, converting molecular nitrogen into nitrites and nitrates that other bacteria, plants, and animals can use. The reactions of nitrogen fixation occur in specialized cells called heterocysts.

Actinobacteria: High G+C Gram-Positive Bacteria
The name Actinobacteria comes from the Greek words for rays and small rod, but Actinobacteria are very diverse. Their microscopic appearance can range from thin filamentous branching rods to coccobacilli. Some Actinobacteria are very large and complex, whereas others are among the smallest independently living organisms. Most Actinobacteria live in the soil, but some are aquatic. The vast majority are aerobic. One distinctive feature of this group is the presence of several different peptidoglycans in the cell wall.
The genus Actinomyces is a much studied representative of Actinobacteria. Actinomyces spp. play an important role in soil ecology, and some species are human pathogens. A number of Actinomyces spp. inhabit the human mouth and are opportunistic pathogens, causing infectious diseases like periodontitis (inflammation of the gums) and oral abscesses. The species A. israelii is an anaerobe notorious for causing endocarditis (inflammation of the inner lining of the heart).The genus Mycobacterium is represented by bacilli covered with a mycolic acid coat. This waxy coat protects the bacteria from some antibiotics, prevents them from drying out, and blocks penetration by Gram stain reagents. Because of this, a special acid-fast staining procedure is used to visualize these bacteria. The genus Mycobacterium is an important cause of a diverse group of infectious diseases. M. tuberculosis is the causative agent of tuberculosis, a disease that primarily impacts the lungs but can infect other parts of the body as well. It has been estimated that one-third of the world’s population has been infected with M. tuberculosis and millions of new infections occur each year. Treatment of M. tuberculosis is challenging and requires patients to take a combination of drugs for an extended time. Complicating treatment even further is the development and spread of multidrug-resistant strains of this pathogen. Another pathogenic species, M. leprae, is the cause of Hansen’s disease (leprosy), a chronic disease that impacts peripheral nerves and the integrity of the skin and mucosal surface of the respiratory tract. Loss of pain sensation and the presence of skin lesions increase susceptibility to secondary injuries and infections with other pathogens.
Low G+C Gram-positive Bacteria Groups
The low G+C gram-positive bacteria have less than 50% guanine and cytosine in their DNA, and this group of bacteria includes a number of genera of bacteria that are pathogenic.
Clostridia
One large and diverse class of low G+C gram-positive bacteria is Clostridia. The best studied genus of this class is Clostridium. These rod-shaped bacteria are generally obligate anaerobes that produce endospores and can be found in anaerobic habitats like soil and aquatic sediments rich in organic nutrients. The endospores may survive for many years.
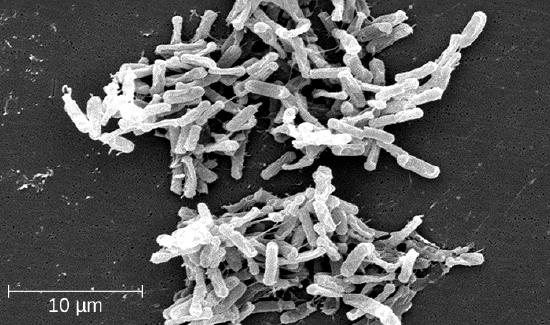
Clostridium spp. produce more kinds of protein toxins than any other bacterial genus, and several species are human pathogens. C. perfringens is the third most common cause of food poisoning in the United States and is the causative agent of an even more serious disease called gas gangrene. Gas gangrene occurs when C. perfringens endospores enter a wound and germinate, becoming viable bacterial cells and producing a toxin that can cause the necrosis (death) of tissue. C. tetani, which causes tetanus, produces a neurotoxin that is able to enter neurons, travel to regions of the central nervous system where it blocks the inhibition of nerve impulses involved in muscle contractions, and cause a life-threatening spastic paralysis. C. botulinum produces botulinum neurotoxin, the most lethal biological toxin known. Botulinum toxin is responsible for rare but frequently fatal cases of botulism. The toxin blocks the release of acetylcholine in neuromuscular junctions, causing flaccid paralysis. In very small concentrations, botulinum toxin has been used to treat muscle pathologies in humans and in a cosmetic procedure to eliminate wrinkles. C. difficile is a common source of hospital-acquired infections (Figure \(\PageIndex{10}\)) that can result in serious and even fatal cases of colitis (inflammation of the large intestine). Infections often occur in patients who are immunosuppressed or undergoing antibiotic therapy that alters the normal microbiota of the gastrointestinal tract.
Lactobacillales
The order Lactobacillales comprises low G+C gram-positive bacteria that include both bacilli and cocci in the genera Lactobacillus, Leuconostoc, Enterococcus, and Streptococcus. Bacteria of the latter three genera typically are spherical or ovoid and often form chains.
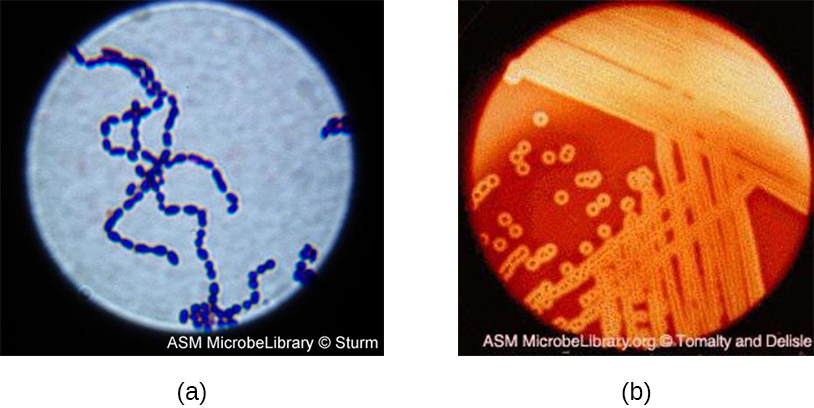
Streptococcus, the name of which comes from the Greek word for twisted chain, is responsible for many types of infectious diseases in humans. Species from this genus, often referred to as streptococci, are usually classified by serotypes called Lancefield groups, and by their ability to lyse red blood cells when grown on blood agar.
S. pyogenes belongs to the Lancefield group A, β-hemolytic Streptococcus. This species is considered a pyogenic pathogen because of the associated pus production observed with infections it causes (Figure \(\PageIndex{11}\)). S. pyogenes is the most common cause of bacterial pharyngitis (strep throat); it is also an important cause of various skin infections that can be relatively mild (e.g., impetigo) or life threatening (e.g., necrotizing fasciitis, also known as flesh eating disease), life threatening.
Bacilli
The name of the class Bacilli suggests that it is made up of bacteria that are bacillus in shape, but it is a morphologically diverse class that includes bacillus-shaped and cocccus-shaped genera. Among the many genera in this class are two that are very important clinically: Bacillus and Staphylococcus. Bacteria in the genus Bacillus are bacillus in shape and can produce endospores. They include aerobes or facultative anaerobes. A number of Bacillus spp. are used in various industries, including the production of antibiotics (e.g., barnase), enzymes (e.g., alpha-amylase, BamH1 restriction endonuclease), and detergents (e.g., subtilisin).
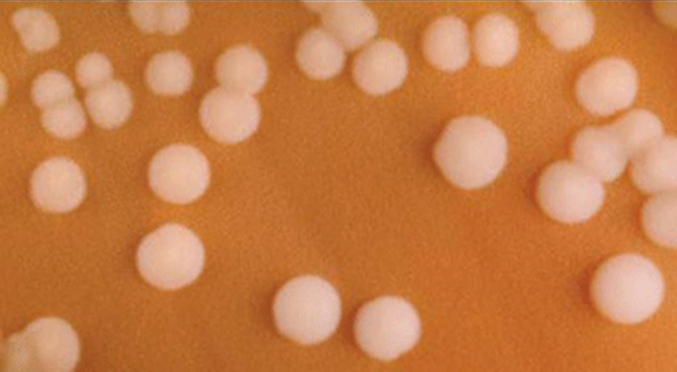
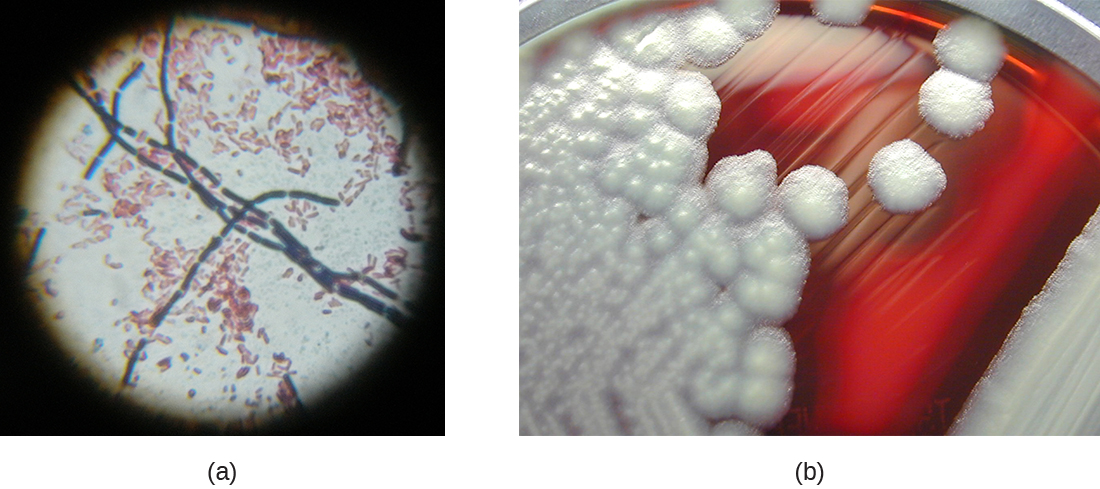
Two notable pathogens belong to the genus Bacillus. B. anthracis is the pathogen that causes anthrax, a severe disease that affects wild and domesticated animals and can spread from infected animals to humans. Anthrax manifests in humans as charcoal-black ulcers on the skin, severe enterocolitis, pneumonia, and brain damage due to swelling. If untreated, anthrax is lethal. B. cereus, a closely related species, is a pathogen that may cause food poisoning. It is a rod-shaped species that forms chains. Colonies appear milky white with irregular shapes when cultured on blood agar (Figure \(\PageIndex{12}\)). One other important species is B. thuringiensis. This bacterium produces a number of substances used as insecticides because they are toxic for insects.
The genus Staphylococcus also belongs to the class Bacilli, even though its shape is coccus rather than a bacillus. The name Staphylococcus comes from a Greek word for bunches of grapes, which describes their microscopic appearance in culture. Staphylococcus spp. are facultative anaerobic, halophilic, and nonmotile. The two best-studied species of this genus are S. epidermidis and S. aureus.
S. epidermidis, whose main habitat is the human skin, is thought to be nonpathogenic for humans with healthy immune systems, but in patients with immunodeficiency, it may cause infections in skin wounds and prostheses (e.g., artificial joints, heart valves). S. epidermidis is also an important cause of infections associated with intravenous catheters. This makes it a dangerous pathogen in hospital settings, where many patients may be immunocompromised.
Strains of S. aureus cause a wide variety of infections in humans, including skin infections that produce boils, carbuncles, cellulitis, or impetigo. Certain strains of S. aureus produce a substance called enterotoxin, which can cause severe enteritis, often called staph food poisoning. Some strains of S. aureus produce the toxin responsible for toxic shock syndrome, which can result in cardiovascular collapse and death.
Mycoplasmas
Although Mycoplasma spp. do not possess a cell wall and, therefore, are not stained by Gram-stain reagents, this genus is still included with the low G+C gram-positive bacteria. The genus Mycoplasma includes more than 100 species, which share several unique characteristics. They are very small cells, some with a diameter of about 0.2 μm, which is smaller than some large viruses. They have no cell walls and, therefore, are pleomorphic, meaning that they may take on a variety of shapes and can even resemble very small animal cells. Because they lack a characteristic shape, they can be difficult to identify. One species, M. pneumoniae, causes the mild form of pneumonia known as “walking pneumonia” or “atypical pneumonia.” This form of pneumonia is typically less severe than forms caused by other bacteria or viruses.
Clinical Focus - Resolution
Marsha’s sputum sample was sent to the microbiology lab to confirm the identity of the microorganism causing her infection. The lab also performed antimicrobial susceptibility testing (AST) on the sample to confirm that the physician has prescribed the correct antimicrobial drugs.
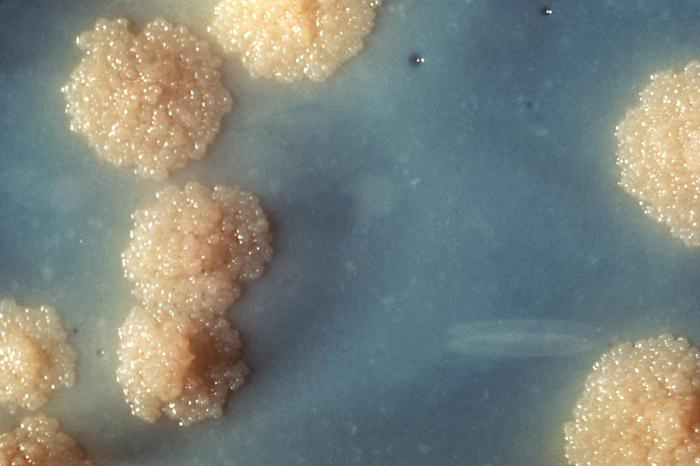
Direct microscopic examination of the sputum revealed acid-fast bacteria (AFB) present in Marsha’s sputum. When placed in culture, there were no signs of growth for the first 8 days, suggesting that microorganism was either dead or growing very slowly. Slow growth is a distinctive characteristic of M. tuberculosis.
After four weeks, the lab microbiologist observed distinctive colorless granulated colonies (Figure \(\PageIndex{13}\)). The colonies contained AFB showing the same microscopic characteristics as those revealed during the direct microscopic examination of Marsha’s sputum. To confirm the identification of the AFB, samples of the colonies were analyzed using nucleic acid hybridization, or direct nucleic acid amplification (NAA) testing. When a bacterium is acid-fast, it is classified in the family Mycobacteriaceae. DNA sequencing of variable genomic regions of the DNA extracted from these bacteria revealed that it was high G+C. This fact served to finalize Marsha’s diagnosis as infection with M. tuberculosis. After nine months of treatment with the drugs prescribed by her doctor, Marsha made a full recovery.
Biopiracy and Bioprospecting
In 1969, an employee of a Swiss pharmaceutical company was vacationing in Norway and decided to collect some soil samples. He took them back to his lab, and the Swiss company subsequently used the fungus Tolypocladium inflatum in those samples to develop cyclosporine A, a drug widely used in patients who undergo tissue or organ transplantation. The Swiss company earns more than $1 billion a year for production of cyclosporine A, yet Norway receives nothing in return—no payment to the government or benefit for the Norwegian people. Despite the fact the cyclosporine A saves numerous lives, many consider the means by which the soil samples were obtained to be an act of “biopiracy,” essentially a form of theft. Do the ends justify the means in a case like this?
Nature is full of as-yet-undiscovered bacteria and other microorganisms that could one day be used to develop new life-saving drugs or treatments.5 Pharmaceutical and biotechnology companies stand to reap huge profits from such discoveries, but ethical questions remain. To whom do biological resources belong? Should companies who invest (and risk) millions of dollars in research and development be required to share revenue or royalties for the right to access biological resources?
Compensation is not the only issue when it comes to bioprospecting. Some communities and cultures are philosophically opposed to bioprospecting, fearing unforeseen consequences of collecting genetic or biological material. Native Hawaiians, for example, are very protective of their unique biological resources.
For many years, it was unclear what rights government agencies, private corporations, and citizens had when it came to collecting samples of microorganisms from public land. Then, in 1993, the Convention on Biological Diversity granted each nation the rights to any genetic and biological material found on their own land. Scientists can no longer collect samples without a prior arrangement with the land owner for compensation. This convention now ensures that companies act ethically in obtaining the samples they use to create their products.
Deeply Branching Bacteria
On a phylogenetic tree, the trunk or root of the tree represents a common ancient evolutionary ancestor, often called the last universal common ancestor (LUCA), and the branches are its evolutionary descendants. Scientists consider the deeply branching bacteria, such as the genus Acetothermus, to be the first of these non-LUCA forms of life produced by evolution some 3.5 billion years ago. When placed on the phylogenetic tree, they stem from the common root of life, deep and close to the LUCA root—hence the name “deeply branching.” (see chapter 1, section 4 for review)
The deeply branching bacteria may provide clues regarding the structure and function of ancient and now extinct forms of life. We can hypothesize that ancient bacteria, like the deeply branching bacteria that still exist, were thermophiles or hyperthermophiles, meaning that they thrived at very high temperatures. Acetothermus paucivorans, a gram-negative anaerobic bacterium discovered in 1988 in sewage sludge, is a thermophile growing at an optimal temperature of 58 °C.6 Scientists have determined it to be the deepest branching bacterium, or the closest evolutionary relative of the LUCA.
The class Thermotogae is represented mostly by hyperthermophilic, as well as some mesophilic (preferring moderate temperatures), anaerobic gram-negative bacteria whose cells are wrapped in a peculiar sheath-like outer membrane called a toga. The thin layer of peptidoglycan in their cell wall has an unusual structure; it contains diaminopimelic acid and D-lysine. These bacteria are able to use a variety of organic substrates and produce molecular hydrogen, which can be used in industry. The class contains several genera, of which the best known is the genus Thermotoga. One species of this genus, T. maritima, lives near the thermal ocean vents and thrives in temperatures of 90 °C; another species, T. subterranea, lives in underground oil reservoirs.
Finally, the deeply branching bacterium Deinococcus radiodurans belongs to a genus whose name is derived from a Greek word meaning terrible berry. Nicknamed “Conan the Bacterium,” D. radiodurans is considered a polyextremophile because of its ability to survive under the many different kinds of extreme conditions—extreme heat, drought, vacuum, acidity, and radiation. It owes its name to its ability to withstand doses of ionizing radiation that kill all other known bacteria; this special ability is attributed to some unique mechanisms of DNA repair.
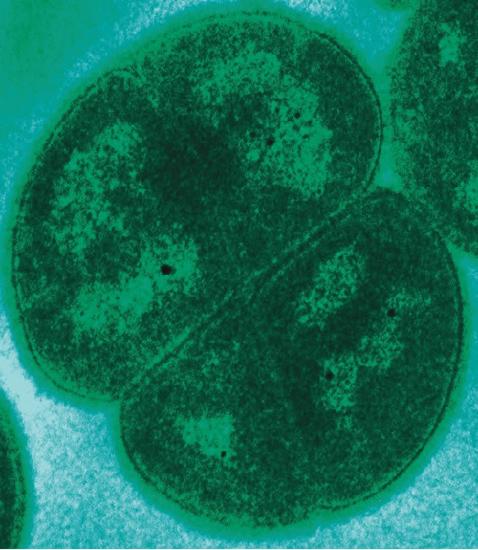
Archaea
Like organisms in the domain Bacteria, organisms of the domain Archaea are all unicellular organisms. However, archaea differ structurally from bacteria in several significant ways. To summarize:
- The archaeal cell membrane is composed of ether linkages with branched isoprene chains (as opposed to the bacterial cell membrane, which has ester linkages with unbranched fatty acids).
- Archaeal cell walls lack peptidoglycan, but some contain a structurally similar substance called pseudopeptidoglycan or pseudomurein.
- The genomes of Archaea are larger and more complex than those of bacteria.
Domain Archaea is as diverse as domain Bacteria, and its representatives can be found in any habitat. Some archaea are mesophiles, and many are extremophiles, preferring extreme hot or cold, extreme salinity, or other conditions that are hostile to most other forms of life on earth. Their metabolism is adapted to the harsh environments, and they can perform methanogenesis, for example, which bacteria and eukaryotes cannot. With few exceptions, archaea are not present in the human microbiota, and none are currently known to be associated with infectious diseases in humans, animals, plants, or microorganisms. However, many play important roles in the environment and may thus have an indirect impact on human health.
The size and complexity of the archaeal genome makes it difficult to classify. Most taxonomists agree that within the Archaea, there are currently five major phyla: Crenarchaeota, Euryarchaeota, Korarchaeota, Nanoarchaeota, and Thaumarchaeota. There are likely many other archaeal groups that have not yet been systematically studied and classified.
Crenarchaeota
Crenarchaeota is a class of Archaea that is extremely diverse, containing genera and species that differ vastly in their morphology and requirements for growth. All Crenarchaeota are aquatic organisms, and they are thought to be the most abundant microorganisms in the oceans. Most, but not all, Crenarchaeota are hyperthermophiles; some of them (notably, the genus Pyrolobus) are able to grow at temperatures up to 113 °C.7
Euryarchaeota
The phylum Euryarchaeota includes several distinct classes. Species in the classes Methanobacteria, Methanococci, and Methanomicrobia represent Archaea that can be generally described as methanogens. Methanogens are unique in that they can reduce carbon dioxide in the presence of hydrogen, producing methane. They can live in the most extreme environments and can reproduce at temperatures varying from below freezing to boiling. Methanogens have been found in hot springs as well as deep under ice in Greenland. Some scientists have even hypothesized that methanogens may inhabit the planet Mars because the mixture of gases produced by methanogens resembles the makeup of the Martian atmosphere.8
Methanogens are thought to contribute to the formation of anoxic sediments by producing hydrogen sulfide, making “marsh gas.” They also produce gases in ruminants and humans. Some genera of methanogens, notably Methanosarcina, can grow and produce methane in the presence of oxygen, although the vast majority are strict anaerobes.
The class Halobacteria (which was named before scientists recognized the distinction between Archaea and Bacteria) includes halophilic (“salt-loving”) archaea. Halobacteria require a very high concentrations of sodium chloride in their aquatic environment. The required concentration is close to saturation, at 36%; such environments include the Dead Sea as well as some salty lakes in Antarctica and south-central Asia. One remarkable feature of these organisms is that they perform photosynthesis using the protein bacteriorhodopsin, which gives them, and the bodies of water they inhabit, a beautiful purple color (Figure \(\PageIndex{15}\)).

Notable species of Halobacteria include Halobacterium salinarum, which may be the oldest living organism on earth; scientists have isolated its DNA from fossils that are 250 million years old.9 Another species, Haloferax volcanii, shows a very sophisticated system of ion exchange, which enables it to balance the concentration of salts at high temperatures
Clinical Focus: Resolution
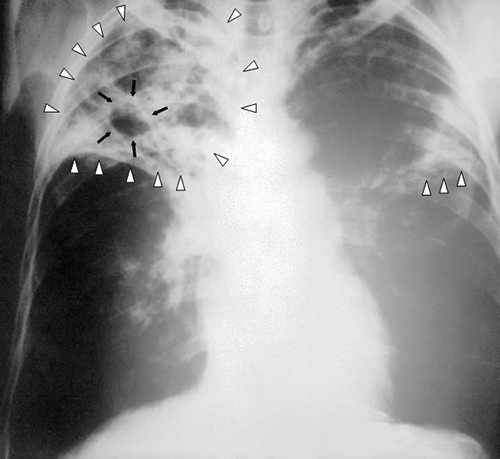
When Marsha finally went to the doctor’s office, the physician listened to her breathing through a stethoscope. He heard some crepitation (a crackling sound) in her lungs, so he ordered a chest radiograph and asked the nurse to collect a sputum sample for microbiological evaluation and cytology. The radiologic evaluation found cavities, opacities, and a particular pattern of distribution of abnormal material (Figure \(\PageIndex{16}\)).
Based on her symptoms, Marsha’s doctor suspected that she had a case of tuberculosis. Although less common in the United States, tuberculosis is still extremely common in many parts of the world, including Nigeria. Marsha’s work there in a medical lab likely exposed her to Mycobacterium tuberculosis, the bacterium that causes tuberculosis.
Marsha’s doctor ordered her to stay at home, wear a respiratory mask, and confine herself to one room as much as possible. He also said that Marsha had to take one semester off school. He prescribed isoniazid and rifampin, antibiotics used in a drug cocktail to treat tuberculosis, which Marsha was to take three times a day for at least three months.
Exercise \(\PageIndex{1}\)
- What are some possible diseases that could be responsible for Marsha’s radiograph results?
- Why did the doctor order Marsha to stay home for three months?
Summary
- In recent years, the traditional approaches to classification of prokaryotes have been supplemented by approaches based on molecular genetics.
- We separate the Phylum in the Bacterial and Archaeal domains by wall composition (Gram positive, Gram negative or other), their G+C percentage, how they get their energy, and how they react to oxygen gas.
- Proteobacteria is a phylum of gram-negative bacteria discovered by Carl Woese in the 1980s based on nucleotide sequence homology. Proteobacteria are further classified into the classes alpha-, beta-, gamma-, delta- and epsilonproteobacteria, each class having separate orders, families, genera, and species.
- Non-proteobacteria that are Gram negative include groups like the spirochetes and photosynthetic bacteria.
- Actinobacteria are a group of high G+C Gram positive bacteria including Actinomyces and Mycobacterium.
- Low G+C Gram positive bacteria include many smaller groups such as Clostridia, Lactobacillales, Bacilli and the highly variable Mycoplasma.
- Deeply branching bacteria are phylogenetically the most ancient forms of life, being the closest to the last universal common ancestor.
- Archaea are unicellular, prokaryotic microorganisms that differ from bacteria in their genetics, biochemistry, and ecology.
- Some archaea are extremophiles, living in environments with extremely high or low temperatures, or extreme salinity.
- Only archaea are known to produce methane. Methane-producing archaea are called methanogens.
Footnotes
- C.R. Woese. “Bacterial Evolution.” Microbiological Review 51 no. 2 (1987):221–271.
- H. Reichenbach. “Myxobacteria, Producers of Novel Bioactive Substances.” Journal of Industrial Microbiology & Biotechnology 27 no. 3 (2001):149–156.
- S. Suerbaum, P. Michetti. “Helicobacter pylori infection.” New England Journal of Medicine 347 no. 15 (2002):1175–1186.
- A. De los Rios et al. “Ultrastructural and Genetic Characteristics of Endolithic Cyanobacterial Biofilms Colonizing Antarctic Granite Rocks.” FEMS Microbiology Ecology 59 no. 2 (2007):386–395.
- J. Andre. Bioethics as Practice. Chapel Hill, NC: University of North Carolina Press, 2002
- G. Dietrich et al. “Acetothermus paucivorans, gen. nov., sp. Nov., a Strictly Anaerobic, Thermophilic Bacterium From Sewage Sludge, Fermenting Hexoses to Acetate, CO2, and H2.” Systematic and Applied Microbiology 10 no. 2 (1988):174–179.
- E. Blochl et al.“Pyrolobus fumani, gen. and sp. nov., represents a novel group of Archaea, extending the upper temperature limit for life to 113°C.” Extremophiles 1 (1997):14–21.
- R.R. Britt “Crater Critters: Where Mars Microbes Might Lurk.” www.space.com/1880-crater-cri...obes-lurk.html. Accessed April 7, 2015.
- H. Vreeland et al. “Fatty acid and DA Analyses of Permian Bacterium Isolated From Ancient Salt Crystals Reveal Differences With Their Modern Relatives.” Extremophiles 10 (2006):71–78.
Contributor
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)


