6.3: Structure and Transcription of RNA
- Page ID
- 31799
Learning Objectives
- Describe the biochemical structure of ribonucleotides
- Describe the similarities and differences between RNA and DNA
- Explain how RNA is synthesized using DNA as a template
- Distinguish between transcription in prokaryotes and eukaryote
- Describe the functions of the three main types of RNA used in protein synthesis
- Explain how RNA can serve as hereditary information
Structurally speaking, ribonucleic acid (RNA), is quite similar to DNA. However, whereas DNA molecules are typically long and double stranded, RNA molecules are much shorter and are typically single stranded. RNA molecules perform a variety of roles in the cell but are mainly involved in the process of protein synthesis (translation) and its regulation.
RNA Structure
RNA is typically single stranded and is made of ribonucleotides that are linked by phosphodiester bonds. A ribonucleotide in the RNA chain contains ribose (the pentose sugar), one of the four nitrogenous bases (A, U, G, and C), and a phosphate group. The subtle structural difference between the sugars gives DNA added stability, making DNA more suitable for storage of genetic information, whereas the relative instability of RNA makes it more suitable for its more short-term functions.
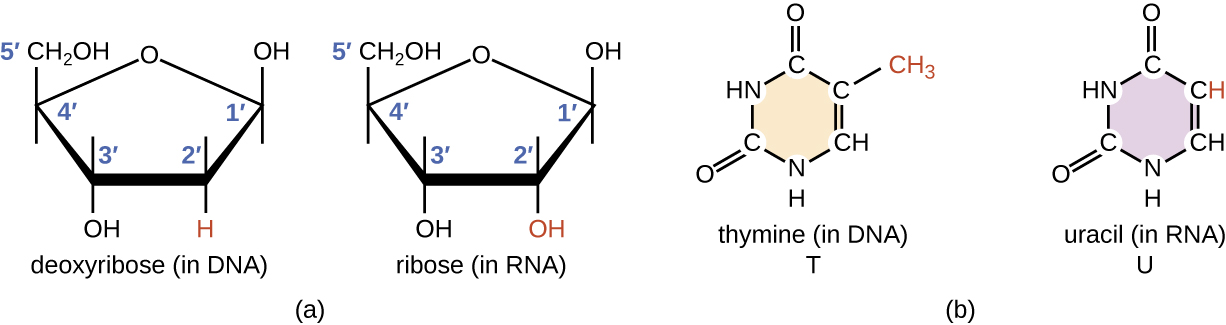
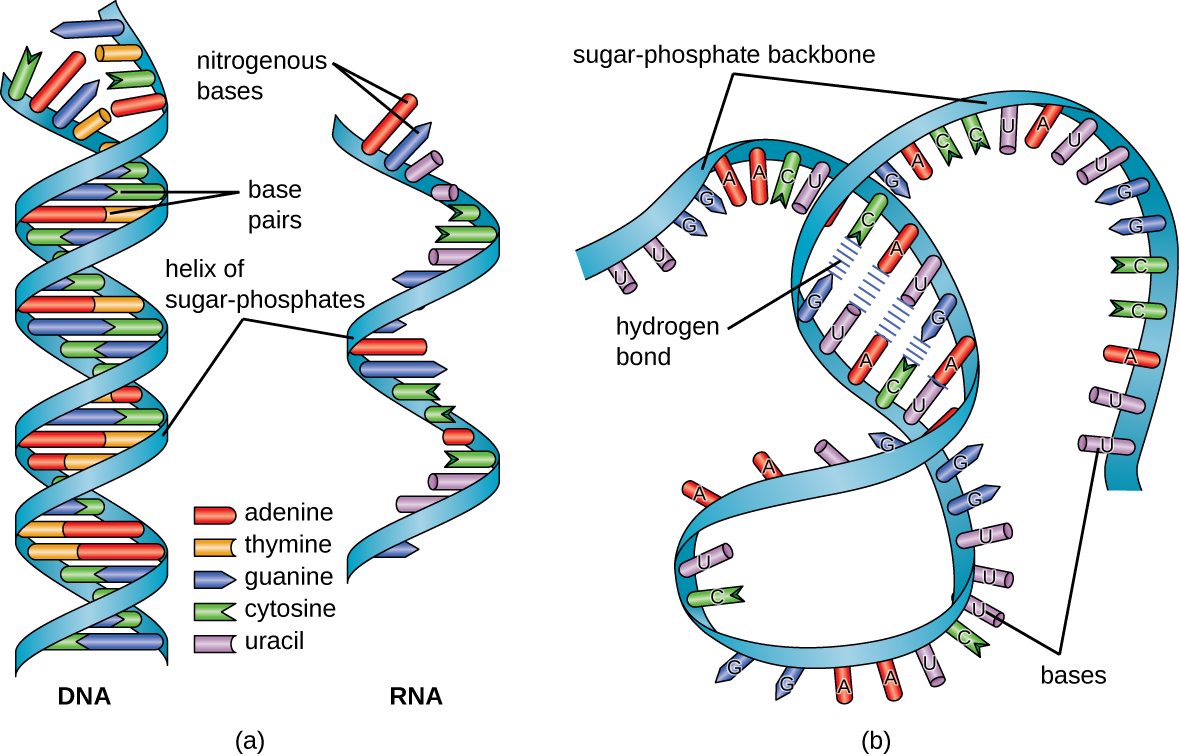
The RNA-specific pyrimidine uracil forms a complementary base pair with adenine and is used instead of the thymine used in DNA. Even though RNA is single stranded, most types of RNA molecules show extensive intramolecular base pairing between complementary sequences within the RNA strand, creating a predictable three-dimensional structure essential for their function (Figure \(\PageIndex{1}\) and Figure \(\PageIndex{2}\)).
Exercise \(\PageIndex{1}\)
How does the structure of RNA differ from the structure of DNA?
Functions of RNA
DNA serves two essential functions that deal with cellular information. First, DNA is the genetic material responsible for inheritance and is passed from parent to offspring for all life on earth. To preserve the integrity of this genetic information, DNA must be replicated with great accuracy, with minimal errors that introduce changes to the DNA sequence. A genome contains the full complement of DNA within a cell and is organized into smaller, discrete units called genes that are arranged on chromosomes and plasmids. The second function of DNA is to direct and regulate the construction of the proteins necessary to a cell for growth and reproduction in a particular cellular environment.
In 1961, French scientists François Jacob and Jacques Monod hypothesized the existence of an intermediary between DNA and its protein products, which they called messenger RNA.1 Evidence supporting their hypothesis was gathered soon afterwards showing that information from DNA is transmitted to the ribosome for protein synthesis using RNA. There are three main types of RNA directly involved in protein synthesis are messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). If DNA serves as the complete library of cellular information, mRNA serves as a photocopy of specific information needed at a particular point in time that serves as the instructions to make a protein. The other types, rRNA and tRNA serve to aid the decoding of mRNA into the correct amino acid sequence which then folds into a protein. Proteins within a cell have many functions, including building cellular structures and serving as enzyme catalysts for cellular chemical reactions that give cells their specific characteristics.
A gene is composed of DNA that is “read” or transcribed to produce an RNA (any type of RNA) molecule during the process of transcription. The processes of transcription and translation are collectively referred to as gene expression. Gene expression is the synthesis of a specific protein with a sequence of amino acids that is encoded in the gene. The flow of genetic information from DNA to RNA to protein is described by the central dogma (Figure \(\PageIndex{3}\)). This central dogma of molecular biology further elucidates the mechanism behind Beadle and Tatum’s “one gene-one enzyme” hypothesis.

Genotype vs Phenotype
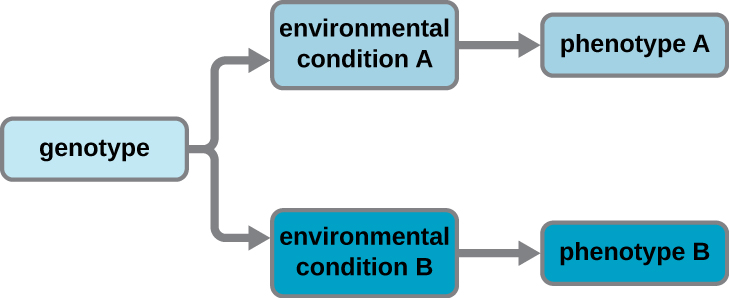
A cell’s genotype is the full collection of genes it contains, whereas its phenotype is the set of observable characteristics that result from those genes. The phenotype is the product of the array of proteins being produced by the cell at a given time, which is influenced by the cell’s genotype as well as interactions with the cell’s environment. Genes code for proteins that have functions in the cell. Production of a specific protein encoded by an individual gene often results in a distinct phenotype for the cell compared with the phenotype without that protein. For this reason, it is also common to refer to the genotype of an individual gene and its phenotype. Although a cell’s genotype remains constant, not all genes are used to direct the production of their proteins simultaneously. Genes that are always expressed are known as constitutive genes; some constitutive genes are known as housekeeping genes because they are necessary for the basic functions of the cell.
Cells carefully regulate expression of most of their genes, only using genes to make specific proteins when those proteins are needed (Figure \(\PageIndex{4}\)). If a cell requires a certain protein to be synthesized, the gene for this product is “turned on” and the mRNA is synthesized through the process of transcription. The mRNA then interacts with ribosomes and other cellular machinery to direct the synthesis of the protein it encodes during the process of translation. The mRNA is relatively unstable and short-lived in the cell, especially in prokaryotic cells, ensuring that proteins are only made when needed.
Exercise \(\PageIndex{2}\)
- What are the two functions of DNA?
- Distinguish between the genotype and phenotype of a cell.
- How can cells have the same genotype but differ in their phenotype?
Transcription
During the process of transcription, the information encoded within the DNA sequence of one or more genes is transcribed into a strand of RNA, also called an RNA transcript. The resulting single-stranded RNA molecule, composed of ribonucleotides containing the bases adenine (A), cytosine (C), guanine (G), and uracil (U), acts as a mobile molecular copy of the original DNA sequence. Transcription in prokaryotes and in eukaryotes requires the DNA double helix to partially unwind in the region of RNA synthesis. The unwound region is called a transcription bubble. Transcription of a particular gene always proceeds from one of the two DNA strands that acts as a template, the so-called antisense strand. The RNA product is complementary to the template strand of DNA and is almost identical to the nontemplate DNA strand, or the sense strand. The only difference is that in RNA, all of the T nucleotides are replaced with U nucleotides; during RNA synthesis, U is incorporated when there is an A in the complementary antisense strand.
Transcription in Bacteria
Bacteria use the same RNA polymerase to transcribe all of their genes. Like DNA polymerase, RNA polymerase adds nucleotides one by one to the 3’-OH group of the growing nucleotide chain. One critical difference in activity between DNA polymerase and RNA polymerase is the requirement for a 3’-OH onto which to add nucleotides: DNA polymerase requires such a 3’-OH group, thus necessitating a primer, whereas RNA polymerase does not. During transcription, a ribonucleotide complementary to the DNA template strand is added to the growing RNA strand and a covalent phosphodiester bond is formed by dehydration synthesis between the new nucleotide and the last one added.
Initiation
The initiation of transcription begins at a promoter, a DNA sequence onto which the transcription machinery binds and initiates transcription. The nucleotide pair in the DNA double helix that corresponds to the site from which the first 5’ RNA nucleotide is transcribed is the initiation site. Nucleotides preceding the initiation site are designated “upstream,” whereas nucleotides following the initiation site are called “downstream” nucleotides. In most cases, promoters are located just upstream of the genes they regulate. Although promoter sequences vary among bacterial genomes, a few elements are conserved. At the –10 and –35 (upstream) positions within the DNA prior to the initiation site (designated +1), there are two promoter consensus sequences, or regions that are similar across all promoters and across various bacterial species. The –10 consensus sequence, called the TATA box, is TATAAT.
Elongation
The elongation in transcription phase begins when the a RNA polymerase subunit dissociates from the polymerase, allowing the core enzyme to synthesize RNA complementary to the DNA template in a 5’ to 3’ direction at a rate of approximately 40 nucleotides per second. As elongation proceeds, the DNA is continuously unwound ahead of the core enzyme and rewound behind it (Figure \(\PageIndex{5}\)).

Termination
Once a gene is transcribed, the bacterial polymerase must dissociate from the DNA template and liberate the newly made RNA. This is referred to as termination of transcription. The DNA template includes repeated nucleotide sequences that act as termination signals, causing RNA polymerase to stall and release from the DNA template, freeing the RNA transcript.
Exercise \(\PageIndex{3}\)
- Where does σ factor of RNA polymerase bind DNA to start transcription?
- What occurs to initiate the polymerization activity of RNA polymerase?
- Where does the signal to end transcription come from?
Transcription in Eukaryotes
Prokaryotes and eukaryotes perform fundamentally the same process of transcription, with a few significant differences. Eukaryotes use three different polymerases, RNA polymerases I, II, and III, all structurally distinct from the bacterial RNA polymerase. Each transcribes a different subset of genes. Interestingly, archaea contain a single RNA polymerase that is more closely related to eukaryotic RNA polymerase II than to its bacterial counterpart. Eukaryotic mRNAs are also usually monocistronic, meaning that they each encode only a single polypeptide, whereas prokaryotic mRNAs of bacteria and archaea are commonly polycistronic, meaning that they encode multiple polypeptides.
The most important difference between prokaryotes and eukaryotes is the latter’s membrane-bound nucleus, which influences the ease of use of RNA molecules for protein synthesis. With the genes bound in a nucleus, the eukaryotic cell must transport protein-encoding RNA molecules to the cytoplasm to be translated. Protein-encoding primary transcripts, the RNA molecules directly synthesized by RNA polymerase, must undergo several processing steps to protect these RNA molecules from degradation during the time they are transferred from the nucleus to the cytoplasm and translated into a protein. For example, eukaryotic mRNAs may last for several hours, whereas the typical prokaryotic mRNA lasts no more than 5 seconds.
The primary transcript (also called pre-mRNA) is first coated with RNA-stabilizing proteins to protect it from degradation while it is processed and exported out of the nucleus. The first type of processing begins while the primary transcript is still being synthesized; a special nucleotide, called the 5’ cap, is added to the 5’ end of the growing transcript. In addition to preventing degradation, factors involved in subsequent protein synthesis recognize the cap, which helps initiate translation by ribosomes. Once elongation is complete, another processing enzyme then adds a string of approximately 200 adenine nucleotides to the 3’ end, called the poly-A tail. This modification further protects the pre-mRNA from degradation and signals to cellular factors that the transcript needs to be exported to the cytoplasm.
Eukaryotic genes that encode polypeptides are composed of coding sequences called exons (ex-on signifies that they are expressed) and intervening sequences called introns (int-ron denotes their intervening role). Transcribed RNA sequences corresponding to introns do not encode regions of the functional polypeptide and are removed from the pre-mRNA during processing. It is essential that all of the intron-encoded RNA sequences are completely and precisely removed from a pre-mRNA before protein synthesis so that the exon-encoded RNA sequences are properly joined together to code for a functional polypeptide. If the process errs by even a single nucleotide, the sequences of the rejoined exons would be shifted, and the resulting polypeptide would be nonfunctional, discussed later in the mutations section. The process of removing intron-encoded RNA sequences and reconnecting those encoded by exons is called RNA splicing and is facilitated by the action of a spliceosome containing small nuclear ribonucleo proteins (snRNPs). Intron-encoded RNA sequences are removed from the pre-mRNA while it is still in the nucleus. Although they are not translated, introns appear to have various functions, including gene regulation and mRNA transport. On completion of these modifications, the mature transcript, the mRNA that encodes a polypeptide, is transported out of the nucleus, destined for the cytoplasm for translation. Introns can be spliced out differently, resulting in various exons being included or excluded from the final mRNA product. This process is known as alternative splicing. The advantage of alternative splicing is that different types of mRNA transcripts can be generated, all derived from the same DNA sequence. In recent years, it has been shown that some archaea also have the ability to splice their pre-mRNA.
Figure \(\PageIndex{6}\): Eukaryotic mRNA contains introns that must be spliced out. A 5' cap and 3' poly-A tail are also added.
Visualize how mRNA splicing happens by watching the process in action in this video. See how introns are removed during RNA splicing here.
Exercise \(\PageIndex{4}\)
- In eukaryotic cells, how is the RNA transcript from a gene for a protein modified after it is transcribed?
- Do exons or introns contain information for protein sequences?
Role of the other RNAs
The two other type of RNA, rRNA and tRNA, are stable types of RNA. In prokaryotes and eukaryotes, tRNA and rRNA are encoded in the DNA, then copied into long RNA molecules that are cut to release smaller fragments containing the individual mature RNA species. In eukaryotes, synthesis, cutting, and assembly of rRNA into ribosomes takes place in the nucleolus region of the nucleus, but these activities occur in the cytoplasm of prokaryotes. Neither of these types of RNA carries instructions to direct the synthesis of a polypeptide, but they play other important roles in protein synthesis.
Ribosomes are composed of rRNA and protein. As its name suggests, rRNA is a major constituent of ribosomes, composing up to about 60% of the ribosome by mass and providing the location where the mRNA binds. The rRNA ensures the proper alignment of the mRNA, tRNA, and the ribosomes; the rRNA of the ribosome also has an enzymatic activity (peptidyl transferase) and catalyzes the formation of the peptide bonds between two aligned amino acids during protein synthesis. Although rRNA had long been thought to serve primarily a structural role, its catalytic role within the ribosome was proven in 2000.2 Scientists in the laboratories of Thomas Steitz (1940–) and Peter Moore(1939–) at Yale University were able to crystallize the ribosome structure from Haloarcula marismortui, a halophilic archaeon isolated from the Dead Sea. Because of the importance of this work, Steitz shared the 2009 Nobel Prize in Chemistry with other scientists who made significant contributions to the understanding of ribosome structure.
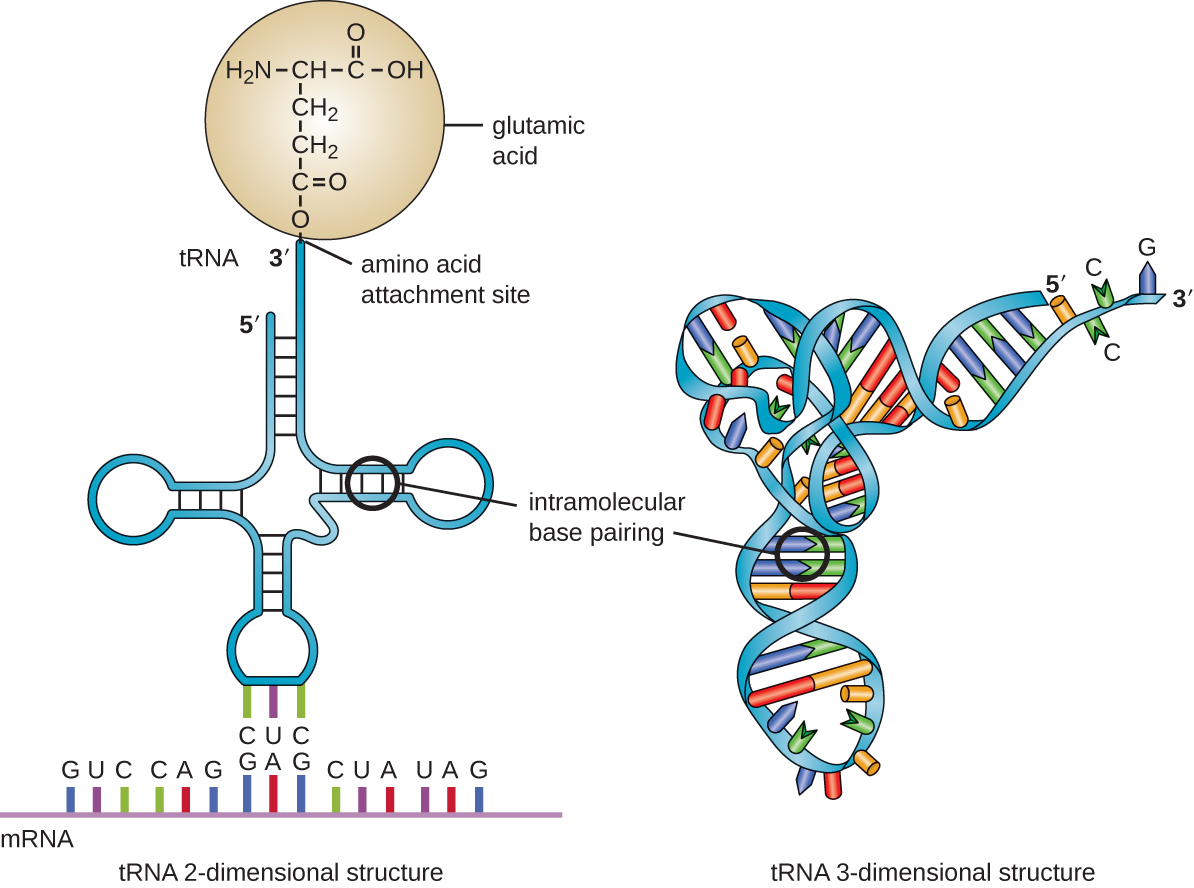
Transfer RNA is the third main type of RNA and one of the smallest, usually only 70–90 nucleotides long. It carries the correct amino acid to the site of protein synthesis in the ribosome. It is the base pairing between the tRNA and mRNA that allows for the correct amino acid to be inserted in the polypeptide chain being synthesized (Figure \(\PageIndex{7}\)). Any mutations in the tRNA or rRNA can result in global problems for the cell because both are necessary for proper protein synthesis (Table \(\PageIndex{1}\)).
| mRNA | rRNA | tRNA | |
|---|---|---|---|
| Structure | Short, unstable, single-stranded RNAcorresponding to a gene encoded within DNA | Longer, stable RNA molecules composing 60% of ribosome’s mass | Short (70-90 nucleotides), stable RNA with extensive intramolecular base pairing; contains an amino acid binding site and an mRNA binding site |
| Function | Serves as intermediary between DNA and protein; used by ribosome to direct synthesis of protein it encodes | Ensures the proper alignment of mRNA, tRNA, and ribosome during protein synthesis; catalyzes peptide bond formation between amino acids | Carries the correct amino acid to the site of protein synthesis in the ribosome |
Exercise \(\PageIndex{5}\)
What are the functions of the three major types of RNA molecules involved in protein synthesis?
RNA as Hereditary Information
Although RNA does not serve as the hereditary information in cells, RNA does hold this function for many viruses that do not contain DNA. Thus, RNA clearly does have the additional capacity to serve as genetic information. Although RNA is typically single stranded within cells, there is significant diversity in viruses. Rhinoviruses, which cause the common cold; influenza viruses; and the Ebola virus are single-stranded RNA viruses. Rotaviruses, which cause severe gastroenteritis in children and other immunocompromised individuals, are examples of double-stranded RNA viruses. Because double-stranded RNA is uncommon in eukaryotic cells, its presence serves as an indicator of viral infection.
Key Concepts and Summary
- Ribonucleic acid (RNA) is typically single stranded and contains ribose as its pentose sugar and the pyrimidine uracil instead of thymine. An RNA strand can undergo significant intramolecular base pairing to take on a three-dimensional structure.
- During transcription, the information encoded in DNA is used to make RNA.
- RNA polymerase synthesizes RNA, using the antisense strand of the DNA as template by adding complementary RNA nucleotides to the 3’ end of the growing strand.
- RNA polymerase binds to DNA at a sequence called a promoter during the initiation of transcription.
- Genes encoding proteins of related functions are frequently transcribed under the control of a single promoter in prokaryotes, resulting in the formation of a polycistronic mRNA molecule that encodes multiple polypeptides.
- Unlike DNA polymerase, RNA polymerase does not require a 3’-OH group to add nucleotides, so a primer is not needed during initiation.
- Termination of transcription in bacteria occurs when the RNA polymerase encounters specific DNA sequences that lead to stalling of the polymerase. This results in release of RNA polymerase from the DNA template strand, freeing the RNA transcript.
- Eukaryotes have three different RNA polymerases. Eukaryotes also have monocistronic mRNA, each encoding only a single polypeptide.
- Eukaryotic primary transcripts are processed in several ways, including the addition of a 5’ cap and a 3′-poly-A tail, as well as splicing, to generate a mature mRNA molecule that can be transported out of the nucleus and that is protected from degradation.
- There are three main types of RNA, all involved in protein synthesis.
- Messenger RNA (mRNA) serves as the intermediary between DNA and the synthesis of protein products during translation.
- Ribosomal RNA (rRNA) is a type of stable RNA that is a major constituent of ribosomes. It ensures the proper alignment of the mRNA and the ribosomes during protein synthesis and catalyzes the formation of the peptide bonds between two aligned amino acids during protein synthesis.
- Transfer RNA (tRNA) is a small type of stable RNA that carries an amino acid to the corresponding site of protein synthesis in the ribosome. It is the base pairing between the tRNA and mRNA that allows for the correct amino acid to be inserted in the polypeptide chain being synthesized.
- Although RNA is not used for long-term genetic information in cells, many viruses do use RNA as their genetic material.
Footnotes
- 1 A. Rich. “The Era of RNA Awakening: Structural Biology of RNA in the Early Years.” Quarterly Reviews of Biophysics 42 no. 2 (2009):117–137.
- 2 P. Nissen et al. “The Structural Basis of Ribosome Activity in Peptide Bond Synthesis.” Science 289 no. 5481 (2000):920–930.
Contributors and Attributions
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)


