1.6: Tools and Media Used for Bacterial Growth
- Page ID
- 31756
Learning Objectives
- Identify and describe tools used in the laboratory setting.
- Describe general protocols for the inoculation, incubation and description of microbes in or on media
- Identify the safety precautions we need to take in a general lab setting.
- Identify and describe culture media for the growth of bacteria, including examples of all-purpose media, enriched, selective, differential, defined, and enrichment media
The study of microorganisms is greatly facilitated if we are able to culture them, that is, to keep reproducing populations alive under laboratory conditions. Culturing many microorganisms is challenging because of highly specific nutritional and environmental requirements and the diversity of these requirements among different species.
Microbiology Toolbox
Because individual microbes are generally too small to be seen with the naked eye, the science of microbiology is dependent on technology that can artificially enhance the capacity of our natural senses of perception. Early microbiologists like Pasteur and Koch had fewer tools at their disposal than are found in modern laboratories, making their discoveries and innovations that much more impressive. Later chapters of this text will explore many applications of technology in depth, but for now, here is a brief overview of some of the fundamental tools of the microbiology lab.
- Growth media are used to grow microorganisms in a lab setting. Some media are liquids; others are more solid or gel-like. A growth medium provides nutrients, including water, various salts, a source of carbon (like glucose), and a source of nitrogen and amino acids (like yeast extract) so microorganisms can grow and reproduce. Ingredients in a growth medium can be modified to grow unique types of microorganisms, or to help separate or identify several microbes grown next to each other.
- Test tubes are cylindrical plastic or glass tubes with rounded bottoms and open tops. They can be used to grow microbes in broth, or semisolid or solid growth media.(Figure \(\PageIndex{1}\)
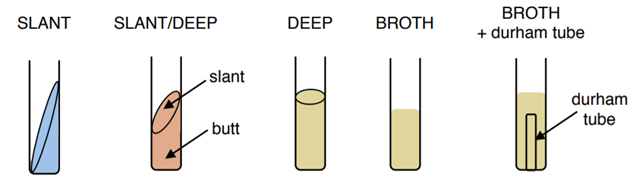
- A Petri dish is a flat-lidded dish that is typically 10–11 centimeters (cm) in diameter and 1–1.5 cm high. Petri dishes made out of either plastic or glass are used to hold thin layers of solid media, usually solidified with agar. (Figure \(\PageIndex{2}\))
- A Bunsen burner is a metal apparatus that creates a flame that can be used to sterilize pieces of equipment. A rubber tube carries gas (fuel) to the burner. In many labs, Bunsen burners are being phased out in favor of infrared microincinerators, which serve a similar purpose without the safety risks of an open flame.
- An inoculation loop is a handheld tool that ends in a small wire loop (Figure \(\PageIndex{2}\)). The loop can be used to streak microorganisms on agar in a Petri dish or to transfer them from one test tube to another. Before each use, the inoculation loop must be sterilized so cultures do not become contaminated, and re-sterilized after the transfer.
- An inoculation needle is another handheld tool that is used for specific solid or semi-solid inoculations. It looks similar to the loop except that it is a straight point. It can help to determine whether an organism is motile (moves). Like the inoculation needle, it needs to be sterilized before every use, and afterwards too.
- Sterile cotton swabs are another common tool of microbiology, useful for collecting samples from surfaces. However they are not able to be reused.
- Microscopes produce magnified images of microorganisms, human cells and tissues, and many other types of specimens too small to be observed with the naked eye. Microscopes will be discussed more in chapter 3.
- Stains and dyes are used to add color to microbes so they can be better observed under a microscope. Some dyes can be used on living microbes, whereas others require that the specimens be fixed with chemicals or heat before staining. Some stains only work on certain types of microbes because of differences in their cellular chemical composition. Staining will be discussed more in chapter 3.
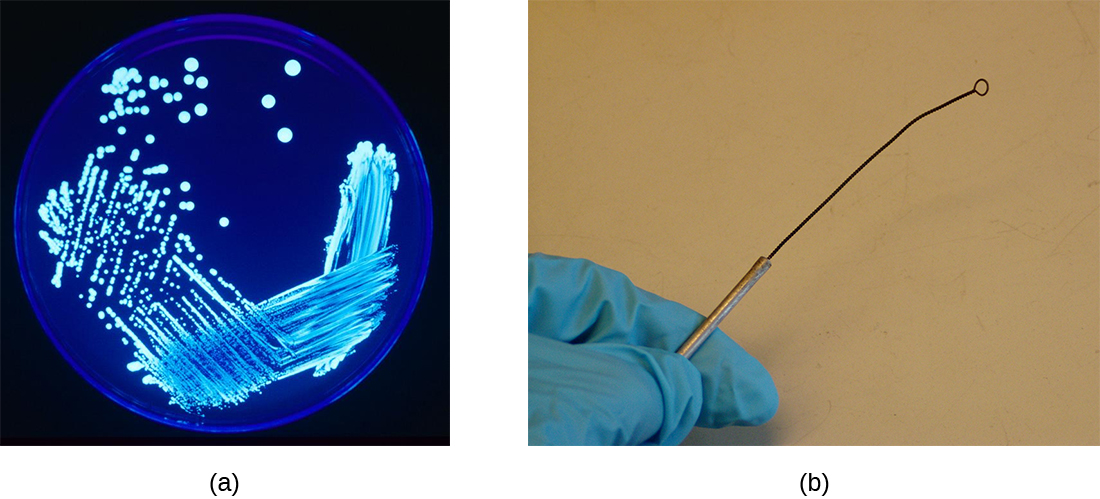
Inoculating and Incubating Media
Aseptic technique is always observed when transferring and inoculating media after applying a label. The inoculation is the introduction of a microbe into "clean" media (media free from other microbes). The instrument used is always sterile or sterilized in a microincincerator or Bunsen Burner first, then used to pick up the sample and applied to the new media. The instrument should either be disposed of or re-sterilized after each inoculation. This is the best procedure to try to ensure a pure culture- a culture with only one microbe growing. Mixed cultures (purposefully adding multiple organisms) are occasionally use to show how isolation streaking can be used to separate organisms. The goal is to avoid a contaminated culture, where unwanted microbes are growing next to or on top of the desired organism.
After the inoculation, the microbe will need to be incubated at its preferred temperature and with its preferred oxygen gas exposure for a number of hours or days. Generally the incubation times are shorter for prokaryotes than for eukaryotes. Test tubes are always incubated in a rack or other holder upright, while plates are incubated upside down.
Describing the Growth of Microbes in Media
Once a microbe has been grown on a plate or in a test tube, they need to be observed for various characteristics. Broth cultures are the easiest to describe as they have the fewest options. Clear broth generally indicates no growth. Cloudy (turbid) broth has growth in it, obscuring the light coming through. Sometimes this growth is only at the top of the tube (pellicle) or only at the bottom (sediment). The last possibility is that the broth has flakes of growth throughout.
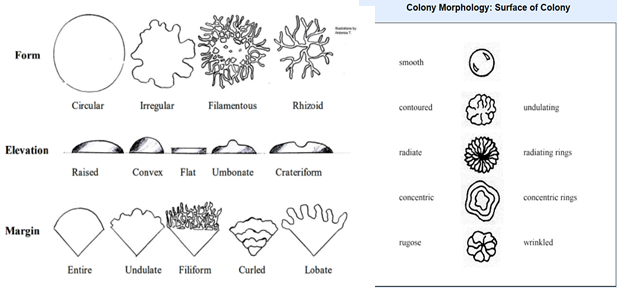
For growth on plates, the technique used to inoculate the plate does make a difference. If the plate was inoculated using a single line inoculation, generally the optical property (opaque, translucent or transparent), overall texture (dry, moist or oily) and the colors of both the microbe and the media are observed. For isolated colonies, especially when identification is desired, more observations are required. When observing isolated colonies we generally describe the following:
- general size (small, medium or large can be used or you can measure its diameter in mm)
- general shape/form (circular, irregular, filamentous, rhizoid)
- optical properties (opaque, translucent or transparent)
- the color of the colony (or colors)
- any discoloration of the agar under or around the colony
- the edge or margin of the colony is described in terms of how smooth it is.
- the surface should be described in terms of both texture (dry, moist or oily) and smoothness of the surface (smooth, contoured, radiating, concentric, rugose)
- elevation as seen from a side-on or profile (raised, convex, flat, umbonate, crateriform)
Laboratory Biological Safety Levels
For researchers or laboratory personnel working with pathogens, the risks associated with specific pathogens determine the levels of cleanliness and control required. The Centers for Disease Control and Prevention (CDC) and the National Institutes of Health (NIH) have established four classification levels, called “biological safety levels” (BSLs). Each BSL requires a different level of biocontainment to prevent contamination and spread of infectious agents to laboratory personnel and, ultimately, the community.
BSL-1 agents are those that generally do not cause infection in healthy human adults. These include noninfectious bacteria, such as nonpathogenic strains of Escherichia coli and Bacillus subtilis, and viruses known to infect bacteria (phages) or animals other than humans, such as baculoviruses (insect viruses). Because working with BSL-1 agents poses very little risk, few precautions are necessary. Laboratory workers use standard aseptic technique and may work with these agents at an open laboratory bench or table, wearing personal protective equipment (PPE) such as a laboratory coat, goggles, and gloves, as needed. Cleaning the area before and after working with a disinfectant like bleach is generally required. Other than a sink for handwashing and a door separating the laboratory from the rest of the building, no additional modifications are needed.
BSL-2 through BSL-4 organism require higher amounts of containment. These will be discussed further in chapter 11.
Handwashing the Right Way
Handwashing is critical for public health and should be emphasized in a clinical setting. For the general public, the CDC recommends handwashing before, during, and after food handling; before eating; before and after interacting with someone who is ill; before and after treating a wound; after using the toilet or changing diapers; after coughing, sneezing, or blowing the nose; after handling garbage; and after interacting with an animal, its feed, or its waste. Figure \(\PageIndex{6}\) illustrates the five steps of proper handwashing recommended by the CDC.
Handwashing is even more important for health-care workers, who should wash their hands thoroughly between every patient contact, after the removal of gloves, after contact with bodily fluids and potentially infectious fomites, and before and after assisting a surgeon with invasive procedures. Even with the use of proper surgical attire, including gloves, scrubbing for surgery is more involved than routine handwashing. The goal of surgical scrubbing is to reduce the normal microbiota on the skin’s surface to prevent the introduction of these microbes into a patient’s surgical wounds.
There is no single widely accepted protocol for surgical scrubbing. Protocols for length of time spent scrubbing may depend on the antimicrobial used; health-care workers should always check the manufacturer’s recommendations. According to the Association of Surgical Technologists (AST), surgical scrubs may be performed with or without the use of brushes (Figure \(\PageIndex{6}\)).
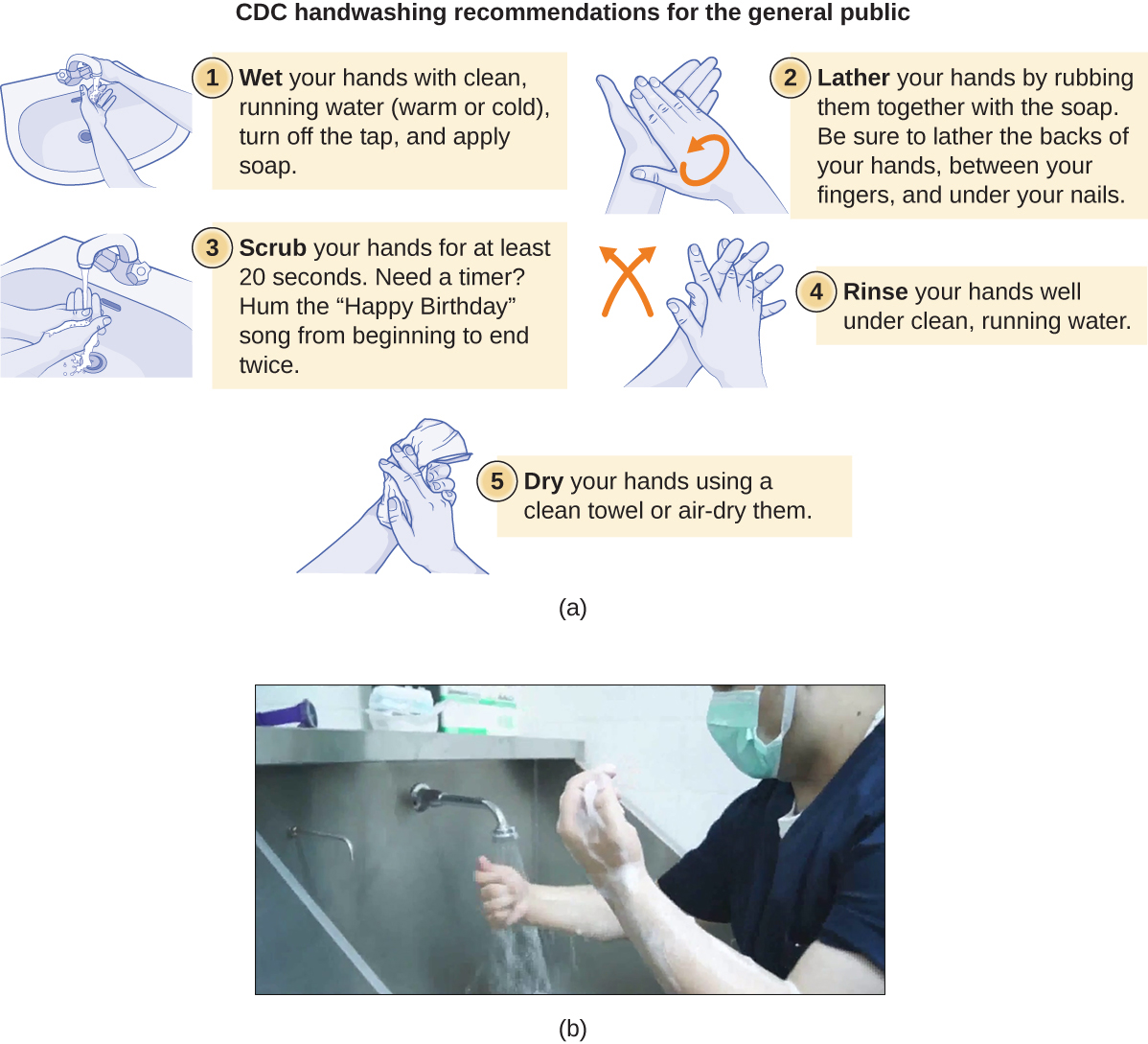
To learn more about proper handwashing, visit the CDC’s website.
Nutritional Requirements
The number of available media to grow bacteria is considerable. Some media are considered general all-purpose media and support growth of a large variety of organisms. A prime example of an all-purpose medium is trypticase soy broth (TSB). Specialized media are used in the identification of bacteria and are supplemented with dyes, pH indicators, or antibiotics. One type, enriched media, contains growth factors, vitamins, and other essential nutrients to promote the growth of fastidious organisms, organisms that cannot make certain nutrients and require them to be added to the medium.
When the complete chemical composition of a medium is known, it is called a chemically defined medium. For example, in EZ medium, all individual chemical components are identified and the exact amounts of each is known. In complex media, which contain extracts and digests of yeasts, meat, or plants, the precise chemical composition of the medium is not known. Amounts of individual components are undetermined and variable. Nutrient broth, such as trypticase soy broth, and brain heart infusion, are all examples of complex media.
Media that inhibit the growth of unwanted microorganisms and support the growth of the organism of interest by supplying nutrients and reducing competition are called selective media. An example of a selective medium is MacConkey agar. It contains bile salts and crystal violet, which interfere with the growth of many gram-positive bacteria and favor the growth of gram-negative bacteria, particularly the Enterobacteriaceae family. The species within this family are commonly named enterics, reside in the intestine, and are adapted to the presence of bile salts. The enrichment cultures foster the preferential growth of a desired microorganism that represents a fraction of the organisms present in an inoculum. For example, if we want to isolate bacteria that break down crude oil, hydrocarbonoclastic bacteria, sequential subculturing (taking a isolated colony and continue to grow it) in a medium that supplies carbon only in the form of crude oil will enrich the cultures with oil-eating bacteria.
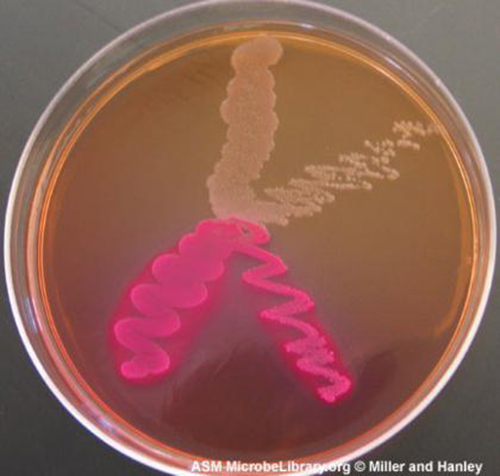
Media that is differential makes it easy to distinguish colonies of different bacteria by a change in the color of the colonies or the color of the medium. Color changes are the result of end products created by interaction of bacterial enzymes with differential substrates in the medium or, in the case of hemolytic reactions, the lysis of red blood cells in the medium. In Figure \(\PageIndex{7}\), the differential fermentation of lactose can be observed on MacConkey agar. The lactose fermenters produce acid, which turns the medium and the colonies of strong fermenters hot pink. The medium is supplemented with the pH indicator neutral red, which turns to hot pink at low pH. Selective and differential media can be combined and play an important role in the identification of bacteria by biochemical methods.
Exercise \(\PageIndex{1}\)
- Distinguish general, specialized and enriched media.
- Distinguish complex and chemically defined media.
- Distinguish selective and differential media.
- Compare the compositions of EZ medium and sheep blood agar.
The End-of-Year Picnic
The microbiology department is celebrating the end of the school year in May by holding its traditional picnic on the green. The speeches drag on for a couple of hours, but finally all the faculty and students can dig into the food: chicken salad, tomatoes, onions, salad, and custard pie. By evening, the whole department, except for two vegetarian students who did not eat the chicken salad, is stricken with nausea, vomiting, retching, and abdominal cramping. Several individuals complain of diarrhea. One patient shows signs of shock (low blood pressure). Blood and stool samples are collected from patients, and an analysis of all foods served at the meal is conducted.
Bacteria can cause gastroenteritis (inflammation of the stomach and intestinal tract) either by colonizing and replicating in the host, which is considered an infection, or by secreting toxins, which is considered intoxication. Signs and symptoms of infections are typically delayed, whereas intoxication manifests within hours, as happened after the picnic.
Blood samples from the patients showed no signs of bacterial infection, which further suggests that this was a case of intoxication. Since intoxication is due to secreted toxins, bacteria are not usually detected in blood or stool samples. MacConkey agar and sorbitol-MacConkey agar plates and xylose-lysine-deoxycholate (XLD) plates were inoculated with stool samples and did not reveal any unusually colored colonies, and no black colonies or white colonies were observed on XLD. All lactose fermenters on MacConkey agar also ferment sorbitol. These results ruled out common agents of food-borne illnesses: E. coli, Salmonella spp., and Shigella spp.
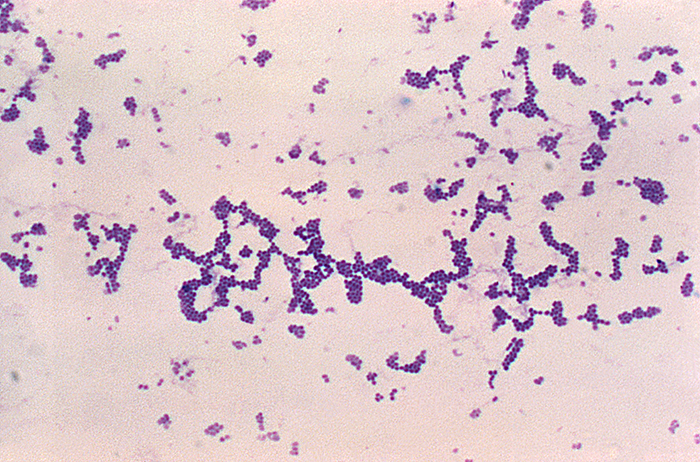
Analysis of the chicken salad revealed an abnormal number of gram-positive cocci arranged in clusters (Figure \(\PageIndex{8}\)). A culture of the gram-positive cocci releases bubbles when mixed with hydrogen peroxide. The culture turned mannitol salt agar yellow after a 24-hour incubation.
All the tests point to Staphylococcus aureus as the organism that secreted the toxin. Samples from the salad showed the presence of gram-positive cocci bacteria in clusters. The colonies were positive for catalase. The bacteria grew on mannitol salt agar fermenting mannitol, as shown by the change to yellow of the medium. The pH indicator in mannitol salt agar is phenol red, which turns to yellow when the medium is acidified by the products of fermentation.
The toxin secreted by S. aureus is known to cause severe gastroenteritis. The organism was probably introduced into the salad during preparation by the food handler and multiplied while the salad was kept in the warm ambient temperature during the speeches.
Exercise \(\PageIndex{2}\)
- What are some other factors that might have contributed to rapid growth of S. aureus in the chicken salad?
- Why would S. aureus not be inhibited by the presence of salt in the chicken salad?
Identifying Bacteria by using API Testing Panels
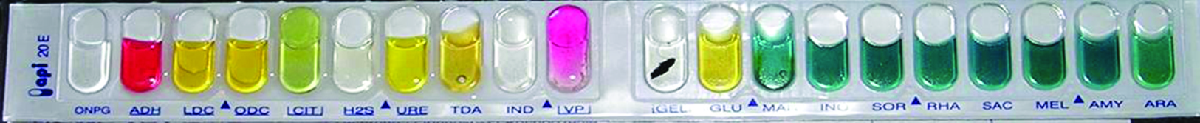
Identification of a microbial isolate is essential for the proper diagnosis and appropriate treatment of patients. Scientists have developed techniques that identify bacteria according to their biochemical characteristics. Typically, they either examine the use of specific carbon sources as substrates for fermentation or other metabolic reactions, or they identify fermentation products or specific enzymes present in reactions. In the past, microbiologists have used individual test tubes and plates to conduct biochemical testing. However, scientists, especially those in clinical laboratories, now more frequently use plastic, disposable, multi-test panels that contain a number of miniature reaction tubes, each typically including a specific substrate and pH indicator. After inoculation of the test panel with a small sample of the microbe in question and incubation, scientists can compare the results to a database that includes the expected results for specific biochemical reactions for known microbes, thus enabling rapid identification of a sample microbe. These test panels have allowed scientists to reduce costs while improving efficiency and reproducibility by performing a larger number of tests simultaneously.
Many commercial, miniaturized biochemical test panels cover a number of clinically important groups of bacteria and yeasts. One of the earliest and most popular test panels is the Analytical Profile Index (API) panel invented in the 1970's. Once some basic laboratory characterization of a given strain has been performed, such as determining the strain’s Gram morphology, an appropriate test strip that contains 10 to 20 different biochemical tests for differentiating strains within that microbial group can be used. Currently, the various API strips can be used to quickly and easily identify more than 600 species of bacteria, both aerobic and anaerobic, and approximately 100 different types of yeasts. Based on the colors of the reactions when metabolic end products are present, due to the presence of pH indicators, a metabolic profile is created from the results (Figure \(\PageIndex{9}\)). Microbiologists can then compare the sample’s profile to the database to identify the specific microbe.
Key Concepts and Summary
- Chemically defined media contain only chemically known components.
- Selective media favor the growth of some microorganisms while inhibiting others.
- Enriched media contain added essential nutrients a specific organism needs to grow
- Differential media help distinguish bacteria by the color of the colonies or the change in the medium.
Footnotes
- Centers for Disease Control and Prevention, World Health Organization. “CDC Laboratory Methods for the Diagnosis of Meningitis Caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenza. WHO Manual, 2nd edition.” 2011. http://www.cdc.gov/meningitis/lab-ma...ull-manual.pdf
Contributors and Attributions
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)


