9.3: Translation
- Page ID
- 35714
What you’ll learn to do: Summarize the process of translation
Take a moment to look at your hands. The bone, skin, and muscle you see are made up of cells. And each of those cells contains many millions of proteins As a matter of fact, proteins are key molecular “building blocks” for every organism on Earth!
How are these proteins made in a cell? For starters, the instructions for making proteins are “written” in a cell’s DNA in the form of genes. Basically, a gene is used to build a protein in a two-step process:
- Step 1: Transcription (which we just learned about)! Here, the DNA sequence of a gene is “rewritten” in the form of RNA. In eukaryotes like you and me, the RNA is processed (and often has a few bits snipped out of it) to make the final product, called a messenger RNA or mRNA.
- Step 2: Translation! In this stage, the mRNA is “decoded” to build a protein (or a chunk/subunit of a protein) that contains a specific series of amino acids.
- Describe the components needed for translation
- Identify the components of the genetic code
- Outline the basic steps of translation
Requirements for Translation
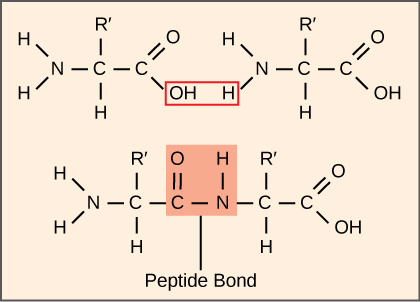
The process of translation, or protein synthesis, involves the decoding of an mRNA message into a polypeptide product. Amino acids are covalently strung together by interlinking peptide bonds. Each individual amino acid has an amino group (NH2) and a carboxyl (COOH) group. Polypeptides are formed when the amino group of one amino acid forms an amide (i.e., peptide) bond with the carboxyl group of another amino acid (Figure 1).
This reaction is catalyzed by ribosomes and generates one water molecule.
The Protein Synthesis Machinery
In addition to the mRNA template, many molecules and macromolecules contribute to the process of translation. Translation requires the input of an mRNA template, ribosomes, tRNAs, and various enzymatic factors.
Click through the steps of this PBS interactive to see protein synthesis in action.
Ribosomes
A ribosome is a complex macromolecule composed of structural and catalytic rRNAs, and many distinct polypeptides. Ribosomes exist in the cytoplasm in prokaryotes and in the cytoplasm and rough endoplasmic reticulum in eukaryotes. Ribosomes are made up of two subunits. In E. coli, the small subunit is described as 30S, and the large subunit is 50S, for a total of 70S. Mammalian ribosomes have a small 40S subunit and a large 60S subunit, for a total of 80S. The small subunit is responsible for binding the mRNA template, whereas the large subunit sequentially binds tRNAs.
tRNAs
The tRNAs are structural RNA molecules that were transcribed from genes by RNA polymerase III. Serving as adaptors, specific tRNAs bind to sequences on the mRNA template and add the corresponding amino acid to the polypeptide chain. Therefore, tRNAs are the molecules that actually “translate” the language of RNA into the language of proteins.

Of the 64 possible mRNA codons—or triplet combinations of A, U, G, and C—three specify the termination of protein synthesis and 61 specify the addition of amino acids to the polypeptide chain. Of these 61, one codon (AUG) also known as the “start codon” encodes the initiation of translation. Each tRNA anticodon can base pair with one of the mRNA codons and add an amino acid or terminate translation, according to the genetic code. For instance, if the sequence CUA occurred on an mRNA template in the proper reading frame, it would bind a tRNA expressing the complementary sequence, GAU, which would be linked to the amino acid leucine.
Mature tRNAs take on a three-dimensional structure through intramolecular hydrogen bonding to position the amino acid binding site at one end and the anticodon at the other end (Figure 2).The anticodon is a three-nucleotide sequence in a tRNA that interacts with an mRNA codon through complementary base pairing.
tRNAs need to interact with three factors:
- They must be recognized by the correct aminoacyl synthetase.
- They must be recognized by ribosomes.
- They must bind to the correct sequence in mRNA.
Aminoacyl tRNA Synthetases
Through the process of tRNA “charging,” each tRNA molecule is linked to its correct amino acid by a group of enzymes called aminoacyl tRNA synthetases. At least one type of aminoacyl tRNA synthetase exists for each of the 20 amino acids.
Genetic Code
Given the different numbers of “letters” in the mRNA and protein “alphabets,” scientists theorized that combinations of nucleotides corresponded to single amino acids. Scientists theorized that amino acids were encoded by nucleotide triplets and that the genetic code was degenerate. In other words, a given amino acid could be encoded by more than one nucleotide triplet. These nucleotide triplets are called codons. Scientists painstakingly solved the genetic code by translating synthetic mRNAs in vitro and sequencing the proteins they specified (Figure 3).

In addition to instructing the addition of a specific amino acid to a polypeptide chain, three (UAA, UAG, UGA) of the 64 codons terminate protein synthesis and release the polypeptide from the translation machinery. These triplets are called nonsense codons, or stop codons. Another codon, AUG, also has a special function. In addition to specifying the amino acid methionine, it also serves as the start codon to initiate translation. The reading frame for translation is set by the AUG start codon near the 5′ end of the mRNA.
The genetic code is universal. With a few exceptions, virtually all species use the same genetic code for protein synthesis. Conservation of codons means that a purified mRNA encoding the globin protein in horses could be transferred to a tulip cell, and the tulip would synthesize horse globin. That there is only one genetic code is powerful evidence that all of life on Earth shares a common origin, especially considering that there are about 1084 possible combinations of 20 amino acids and 64 triplet codons.
Transcribe a gene and translate it to protein using complementary pairing and the genetic code at this site.
Degeneracy is believed to be a cellular mechanism to reduce the negative impact of random mutations. Codons that specify the same amino acid typically only differ by one nucleotide. In addition, amino acids with chemically similar side chains are encoded by similar codons. This nuance of the genetic code ensures that a single-nucleotide substitution mutation might either specify the same amino acid but have no effect or specify a similar amino acid, preventing the protein from being rendered completely nonfunctional.
Steps of Translation
As with mRNA synthesis, protein synthesis can be divided into three phases: initiation, elongation, and termination. The process of translation is similar in prokaryotes and eukaryotes. Here we’ll explore how translation occurs in E. coli, a representative prokaryote, and specify any differences between prokaryotic and eukaryotic translation.
Initiation of Translation
Protein synthesis begins with the formation of an initiation complex. In E. coli, this complex involves the small 30S ribosome, the mRNA template, initiation factors and a special initiator tRNA. The initiator tRNA interacts with the start codon AUG. Guanosine triphosphate (GTP), which is a purine nucleotide triphosphate, acts as an energy source during translation—both at the start of elongation and during the ribosome’s translocation.
Once the appropriate AUG is identified, the 50S subunit binds to the complex of Met-tRNAi, mRNA, and the 30S subunit. This step completes the initiation of translation.
Elongation of Translation
The 50S ribosomal subunit of E. coli consists of three compartments: the A (aminoacyl) site binds incoming charged aminoacyl tRNAs. The P (peptidyl) site binds charged tRNAs carrying amino acids that have formed peptide bonds with the growing polypeptide chain but have not yet dissociated from their corresponding tRNA. The E (exit) site releases dissociated tRNAs so that they can be recharged with free amino acids. this creates an initiation complex with a free A site ready to accept the tRNA corresponding to the first codon after the AUG.

During translation elongation, the mRNA template provides specificity. As the ribosome moves along the mRNA, each mRNA codon comes into register, and specific binding with the corresponding charged tRNA anticodon is ensured. If mRNA were not present in the elongation complex, the ribosome would bind tRNAs nonspecifically.
Elongation proceeds with charged tRNAs entering the A site and then shifting to the P site followed by the E site with each single-codon “step” of the ribosome. Ribosomal steps are induced by conformational changes that advance the ribosome by three bases in the 3′ direction. The energy for each step of the ribosome is donated by an elongation factor that hydrolyzes GTP. Peptide bonds form between the amino group of the amino acid attached to the A-site tRNA and the carboxyl group of the amino acid attached to the P-site tRNA. The formation of each peptide bond is catalyzed by peptidyl transferase, an RNA-based enzyme that is integrated into the 50S ribosomal subunit. The energy for each peptide bond formation is derived from GTP hydrolysis, which is catalyzed by a separate elongation factor. The amino acid bound to the P-site tRNA is also linked to the growing polypeptide chain. As the ribosome steps across the mRNA, the former P-site tRNA enters the E site, detaches from the amino acid, and is expelled (Figure 5). Amazingly, the E. coli translation apparatus takes only 0.05 seconds to add each amino acid, meaning that a 200-amino acid protein can be translated in just 10 seconds.

Many antibiotics inhibit bacterial protein synthesis. For example, tetracycline blocks the A site on the bacterial ribosome, and chloramphenicol blocks peptidyl transfer. What specific effect would you expect each of these antibiotics to have on protein synthesis?
Tetracycline would directly affect:
- tRNA binding to the ribosome
- ribosome assembly
- growth of the protein chain
- Show Answer
-
Answer a. Tetracycline would directly affect tRNA binding to the ribosome.
Chloramphenicol would directly affect
- tRNA binding to the ribosome
- ribosome assembly
- growth of the protein chain
- Show Answer
-
Answer c. Chloramphenicol would directly affect growth of the protein chain.
Termination of Translation
Termination of translation occurs when a nonsense codon (UAA, UAG, or UGA) is encountered. Upon aligning with the A site, these nonsense codons are recognized by release factors in prokaryotes and eukaryotes that instruct peptidyl transferase to add a water molecule to the carboxyl end of the P-site amino acid. This reaction forces the P-site amino acid to detach from its tRNA, and the newly made protein is released. The small and large ribosomal subunits dissociate from the mRNA and from each other; they are recruited almost immediately into another translation initiation complex. After many ribosomes have completed translation, the mRNA is degraded so the nucleotides can be reused in another transcription reaction.
Check Your Understanding
Answer the question(s) below to see how well you understand the topics covered in the previous section. This short quiz does not count toward your grade in the class, and you can retake it an unlimited number of times.
Use this quiz to check your understanding and decide whether to (1) study the previous section further or (2) move on to the next section.
Contributors and Attributions
- Introduction to Translation. Authored by: Shelli Carter and Lumen Learning. Provided by: Lumen Learning. License: CC BY: Attribution
- Overview of translation. Provided by: Khan Academy. Located at: https://www.khanacademy.org/science/biology/gene-expression-central-dogma/translation-polypeptides/a/translation-overview. License: CC BY-NC-SA: Attribution-NonCommercial-ShareAlike
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72...f21b5eabd@10.8

