13.3: Expression of Genes
- Page ID
- 43706
For a cell to function properly, necessary proteins must be synthesized at the proper time. All cells control or regulate the synthesis of proteins from information encoded in their DNA. The process of turning on a gene to produce RNA and protein is called gene expression. Whether in a simple unicellular organism or a complex multi-cellular organism, each cell controls when and how its genes are expressed. For this to occur, there must be a mechanism to control when a gene is expressed to make RNA and protein, how much of the protein is made, and when it is time to stop making that protein because it is no longer needed.
The regulation of gene expression conserves energy and space. It would require a significant amount of energy for an organism to express every gene at all times, so it is more energy efficient to turn on the genes only when they are required. In addition, only expressing a subset of genes in each cell saves space because DNA must be unwound from its tightly coiled structure to transcribe and translate the DNA. Cells would have to be enormous if every protein were expressed in every cell all the time.
The control of gene expression is extremely complex. Malfunctions in this process are detrimental to the cell and can lead to the development of many diseases, including cancer.
Gene regulation makes cells different
Gene regulation is how a cell controls which genes, out of the many genes in its genome, are “turned on” (expressed). Thanks to gene regulation, each cell type in your body has a different set of active genes—despite the fact that almost all the cells of your body contain the exact same DNA. These different patterns of gene expression cause your various cell types to have different sets of proteins, making each cell type uniquely specialized to do its job.
For example, one of the jobs of the liver is to remove toxic substances like alcohol from the bloodstream. To do this, liver cells express genes encoding subunits (pieces) of an enzyme called alcohol dehydrogenase. This enzyme breaks alcohol down into a non-toxic molecule. The neurons in a person’s brain don’t remove toxins from the body, so they keep these genes unexpressed, or “turned off.” Similarly, the cells of the liver don’t send signals using neurotransmitters, so they keep neurotransmitter genes turned off (Figure 1).
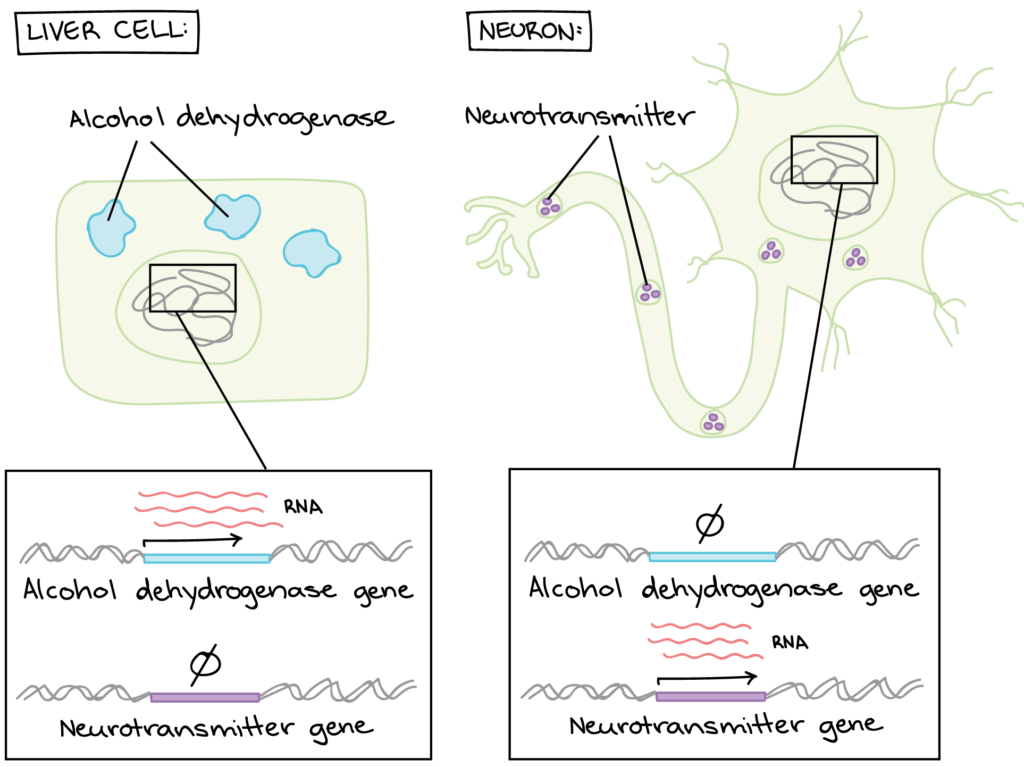
There are many other genes that are expressed differently between liver cells and neurons (or any two cell types in a multicellular organism like yourself).
How do cells “decide” which genes to turn on?
Now there’s a tricky question! Many factors that can affect which genes a cell expresses. Different cell types express different sets of genes, as we saw above. However, two different cells of the same type may also have different gene expression patterns depending on their environment and internal state.
Broadly speaking, we can say that a cell’s gene expression pattern is determined by information from both inside and outside the cell.
- Examples of information from inside the cell: the proteins it inherited from its mother cell, whether its DNA is damaged, and how much ATP it has.
- Examples of information from outside the cell: chemical signals from other cells, mechanical signals from the extracellular matrix, and nutrient levels.
How do these cues help a cell “decide” what genes to express? Cells don’t make decisions in the sense that you or I would. Instead, they have molecular pathways that convert information—such as the binding of a chemical signal to its receptor—into a change in gene expression.
As an example, let’s consider how cells respond to growth factors. A growth factor is a chemical signal from a neighboring cell that instructs a target cell to grow and divide. We could say that the cell “notices” the growth factor and “decides” to divide, but how do these processes actually occur?
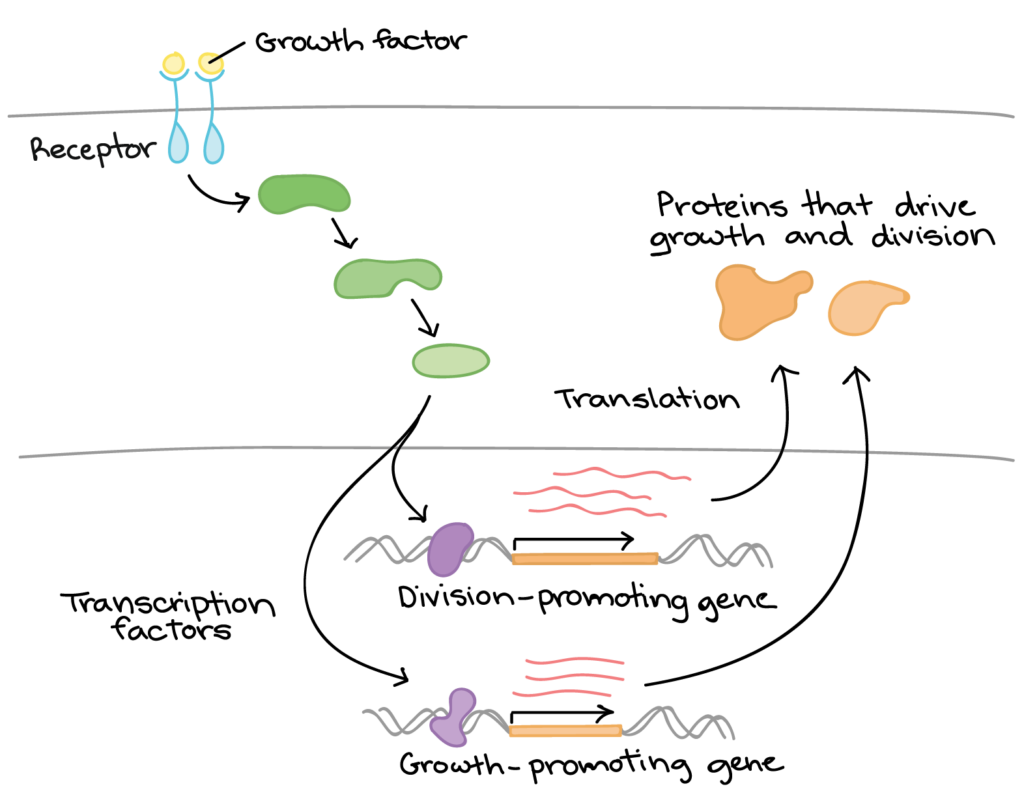
- The cell detects the growth factor through physical binding of the growth factor to a receptor protein on the cell surface.
- Binding of the growth factor causes the receptor to change shape, triggering a series of chemical events in the cell that activate proteins called transcription factors.
- The transcription factors bind to certain sequences of DNA in the nucleus and cause transcription of cell division-related genes.
- The products of these genes are various types of proteins that make the cell divide (drive cell growth and/or push the cell forward in the cell cycle).
This is just one example of how a cell can convert a source of information into a change in gene expression. There are many others, and understanding the logic of gene regulation is an area of ongoing research in biology today.
Growth factor signaling is complex and involves the activation of a variety of targets, including both transcription factors and non-transcription factor proteins.
Learning Objectives
- Gene regulation is the process of controlling which genes in a cell’s DNA are expressed (used to make a functional product such as a protein).
- Different cells in a multicellular organism may express very different sets of genes, even though they contain the same DNA.
- The set of genes expressed in a cell determines the set of proteins and functional RNAs it contains, giving it its unique properties.
- In eukaryotes like humans, gene expression involves many steps, and gene regulation can occur at any of these steps. However, many genes are regulated primarily at the level of transcription.
[reveal-answer q=”915423″]Show References[/reveal-answer]
[hidden-answer a=”915423″]
Alcohol dehydrogenase. (2016, January 6). Retrieved April 26, 2016 from Wikipedia: https://en.Wikipedia.org/wiki/Alcohol_dehydrogenase.
Cooper, G. M. (2000). Regulation of transcription in eukaryotes. In The cell: A molecular approach. Sunderland, MA: Sinauer Associates. Retrieved from www.ncbi.nlm.nih.gov/books/NBK9904/.
Kimball, John W. (2014, April 19). The human and chimpanzee genomes. In Kimball’s biology pages. Retrieved from http://www.biology-pages.info/H/HominoidClade.html.
OpenStax College, Biology. (2016, March 23). Eukaryotic transcription gene regulation. In _OpenStax CNX. Retrieved from http://cnx.org/contents/GFy_h8cu@10....cription-Gene-.
OpenStax College, Biology. (2016, March 23). Regulation of gene expression. In _OpenStax CNX. Retrieved from http://cnx.org/contents/GFy_h8cu@10....ene-Expression
Phillips, T. (2008). Regulation of transcription and gene expression in eukaryotes. Nature Education, 1(1), 199. Retrieved from http://www.nature.com/scitable/topic...ession-in-1086.
Purves, W. K., Sadava, D. E., Orians, G. H., and Heller, H.C. (2003). Transcriptional regulation of gene expression. In Life: The science of biology (7th ed., pp. 290-296). Sunderland, MA: Sinauer Associates.
Reece, J. B., Urry, L. A., Cain, M. L., Wasserman, S. A., Minorsky, P. V., and Jackson, R. B. (2011). Eukaryotic gene expression is regulated at many stages. In Campbell Biology (10th ed., pp. 365-373). San Francisco, CA: Pearson.
[/hidden-answer]
Contributors and Attributions
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72...f21b5eabd@10.8
- Overview: Eukaryotic gene regulation. Provided by: Khan Academy. Located at: https://www.khanacademy.org/science/biology/gene-regulation/gene-regulation-in-eukaryotes/a/overview-of-eukaryotic-gene-regulation. License: CC BY-NC-SA: Attribution-NonCommercial-ShareAlike

