12.6: RNA Post-Translational Modification
- Page ID
- 43693
Learning Objectives
- Gene expression can be regulated at various stages after an RNA transcript has been produced.
- Some transcripts can undergo alternative splicing. This regulated process makes different mRNAs and proteins from the same initial RNA transcript.
- Some mRNAs are targeted by small regulatory RNAs, including miRNAs, which can cause mRNA degradation or block translation.
- A protein’s activity may be regulated after translation by mechanisms such as proteolysis (“snipping out” of pieces) and addition of chemical groups.
The genes that a eukaryotic cell turns “on” largely determine its identity and properties. For instance, a photoreceptor cell in your eye can detect light because it expresses genes for light-sensitive proteins, as well as as genes for neurotransmitters that allow signals to be relayed to the brain.
In eukaryotic cells like photoreceptors, gene expression is often controlled primarily at the level of transcription. However, that doesn’t mean transcription is the last chance for regulation. Later stages of gene expression can also be regulated, including the following:
- RNA processing, such as splicing, capping, and addition of a poly-A tail
- Messenger RNA (mRNA) translation and lifetime in the cytosol
- Protein modifications, such as addition of chemical groups or removal of amino acids
In the sections below, we’ll discuss some common types of gene regulation that occur after an RNA transcript has been made.
Regulation of RNA processing
When a eukaryotic gene is transcribed in the nucleus, the primary transcript (freshly made RNA molecule) isn’t yet considered a messenger RNA. Instead, it’s an “immature” molecule called a pre-mRNA.
The pre-mRNA has to go through some modifications to become a mature mRNA molecule that can leave the nucleus and be translated. These include splicing, capping, and addition of a poly-A tail, all of which can potentially be regulated – sped up, slowed down, or altered to result in a different product.
Alternative splicing
Most pre-mRNA molecules have sections that are removed from the molecule, called introns, and sections that are linked or together to make the final mRNA, called exons. This process is called splicing.
In the process of alternative splicing, different portions of an mRNA can be selected for use as exons. This allows either of two (or more) mRNA molecules to be made from one pre-mRNA.
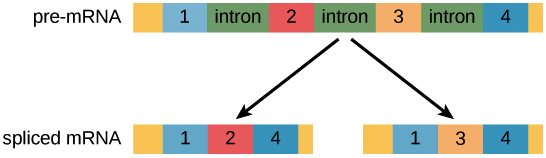
Alternative splicing is not a random process. Instead, it’s typically controlled by regulatory proteins. The proteins bind to specific sites on the pre-mRNA and “tell” the splicing factors which exons should be used. Different cell types may express different regulatory proteins, so different exon combinations can be used in each cell type, leading to the production of different proteins.
Small regulatory RNA
Once an mRNA has left the nucleus, it may or may not be translated many times to make proteins. Two key determinants of how much protein is made from an mRNA are its “lifespan” (how long it floats around in the cytosol) and how readily the translation machinery, such as the ribosome, can attach to it.
A recently discovered class of regulators, called small regulatory RNAs, can control mRNA lifespan and translation. Let’s see how this works.
microRNAs
microRNAs (miRNAs) were among the first small regulatory RNAs to be discovered. A miRNA is first transcribed as a long RNA molecule, which forms base pairs with itself and folds over to make a hairpin. Next, the hairpin is chopped up by enzymes, releasing a small double-stranded fragment of about 20 nucleotides. One of the strands in this fragment is the mature miRNA, which binds to a specific protein to make an RNA-protein complex.

The miRNA directs the protein complex to “matching” mRNA molecules (ones that form base pairs with the miRNA). When the RNA-protein complex binds:
- If the miRNA and its target match perfectly, an enzyme in the RNA-protein complex will typically chop the mRNA in half, leading to its breakdown.
- If the miRNA and its target have some mismatches, the RNA-protein complex may instead bind to the mRNA and keep it from being translated.
These are not the only ways that miRNAs inhibit expression of their targets, and scientists are still investigating their many modes of action.[1]
- Carthew, R. W. and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell, 136(4), 642–655. [1]http://dx.doi.org/10.1016/j.cell.2009.01.035. ↵
Contributors and Attributions
- Regulation after transcription. Provided by: Khan Academy. Located at: https://www.khanacademy.org/science/biology/gene-regulation/gene-regulation-in-eukaryotes/a/regulation-after-transcription. License: CC BY-NC-SA: Attribution-NonCommercial-ShareAlike

