16.2: Introduction
- Page ID
- 40254
This lab will demonstrate principles of
- Bacterial reproduction
- Genetic recombination in bacteria
- Transformation as a method of genetic exchange and recombination
- Transformation and genetic exchange as a vehicle of antibiotic resistance
- Gene regulation in bacteria
- Genetic engineering and recombinant organisms
Fred Griffith, a British microbiologist, discovered in 1928 that virulence could be transferred from killed pathogenic bacteria to live non-pathogenic bacteria.2 He called this process “transformation”. In 1944, Oswald Avery and his research group discovered that DNA was the factor being transferred.3 Thesescientists had discovered and defined transformation, one of the mechanisms of bacterial recombination (Joshua Lederberg and others discovered the other two methods, conjugation and transduction). This was also the beginning of molecular biology and genetic engineering.
Today we know that genetic recombination in bacteria can be a source of antibiotic resistance. In addition, genes that code for antibiotic resistance mechanisms—like enzymes that break down the antibiotic in the bacterium—are carried on plasmids.
In this lab you will demonstrate the processes of transformation and gene regulation in bacteria, and genetic engineering, by forcing a bacterium to take up plasmid DNA from its environment (transformation) and express a gene (gene regulation) it would not normally have (genetic engineering). Typically, a very low percentage of bacterial cells will actually be transformed during this process. Some will successfully take up the plasmid; others may reject the plasmid in one of a few ways. But remember, most bacteria can grow so fast that, even if only one in a million are transformed with a plasmid that gives the cell resistance to an antibiotic, one cell can be millions in a matter of hours!
When I was a grad student at UCLA, I engineered a custom recombinant plasmid to express genes that I had mutated. The plasmid had the same amp R gene as pAMP and pGLO, so I was able to select for transformants by plating the cells on media that contained ampicillin. After inserting the mutated gene into the plasmid, I transformed E. coli cells and conducted growth experiments. My plasmid eventually became very popular with other researchers in the Microbiology, Immunology and Molecular Genetics Department, who nicknamed it ‘pUC-Shane’!
Dr. C. Shane Ramey
There are two sides to the process in the lab you will do. First of all, you will insert a plasmid (called pAMP) that contains a gene (ampR) that confers resistance to the antibiotic Ampicillin into E. coli. Therefore, the E. coli is transformed and if subsequently exposed to Ampicillin it will not be affected and will grow as usual. Transformation happens in nature and is a vehicle for the spread of antibiotic resistance in bacteria. In a lab, antibiotic resistance genes can be paired with other genes on the same plasmid. These are called recombinant plasmids because they have been constructed from genes of different organisms. One could insert a plasmid that contains a specific gene for a desirable protein. But, it might be difficult to know which bacteria have successfully taken up the plasmid and the gene(s) it contains. To be efficient, one would want to select the bacteria that can make protein (called transformants--the bacteria that have been successfully transformed), and not end up with bacteria that cannot. Therefore, if an antibiotic resistance gene is paired with the other gene (the gene of interest), then the acquisition of resistance, which is easily tested, indicates the acquisition of the gene of interest. The cells resistant to Ampicillin are then “selectable markers” for the other gene.
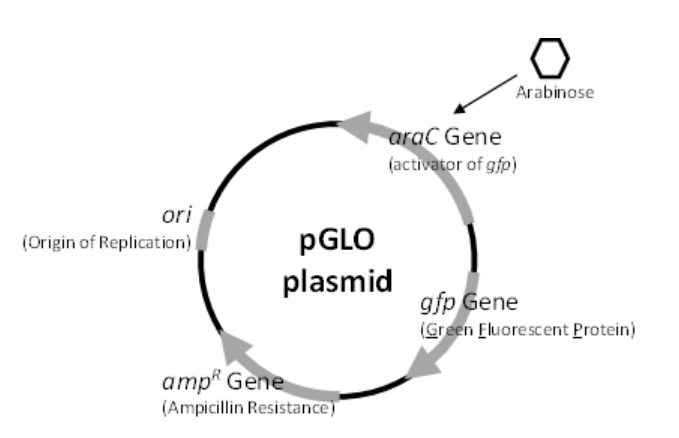
The second aspect of today’s process, then, is that the plasmid with Ampicillin resistance also carries the gene (the gene of interest) to produce green fluorescent protein (called pGLO which has the AMPR andgfp genes-see Figure \(\PageIndex{1}\)), which glows when exposed to UV light. Cells that have pGLO will fluoresce only when under the correct environmental conditions, specifically the presence of the sugar Arabinose. You will use the antibiotic resistance property to select for cells that, potentially, will fluoresce. Then, we’ll feed the transformed bacteria Arabinose in order to “turn on” the genes for fluorescence. The presence of the Arabinose sugar is the environmental switch that helps regulate the expression/production of the green fluorescent protein. As you will observe in today’s lab, bacteria plated onto media that contains Ampicillin will grow, and if the media also contains Arabinose, they will glow! You will have successfully selected for a bacterium that you actually engineered to fluoresce.
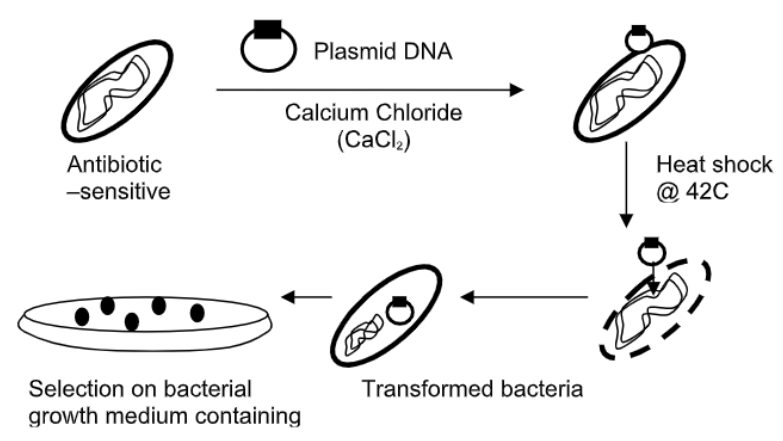
In nature, not all bacterial cells can undergo transformation. Those that are able are called “competent” cells. In the laboratory, cells can be encouraged to undergo transformation by altering their environment and cells temporarily so that they will uptake DNA; they become competent due to lab manipulation. The method you will use is the calcium chloride/ heat shock procedure (see Figure \(\PageIndex{2}\)). The positive Ca ions released in the solution will neutralize the negative charge on the DNA molecule (a plasmid in our case), which reduces the charge barrier for the DNA entering the cell (remember that cells have a net negative charge). Heating the cells increases the permeability of the bacterial membrane. Thus, DNA will more readily enter the bacterial cells and transformation may proceed. These steps must be done quickly in order not to damage or kill the cells. Later, you will calculate the “Transformation Efficiency” obtained by your bacterial culture. Not all cells will become competent, nor will all competent cells complete transformation.
Contributors and Attributions
Kelly C. Burke (College of the Canyons)


