3.3: Enzyme Regulation
- Page ID
- 3152
- Compare and contrast the genetic control of enzyme activity (enzyme synthesis) in bacteria with the control of enzyme activity through feedback inhibition.
- Compare and contrast an inducible operon with a repressible operon and give an example of each.
- Compare how the presense or absence of tryptophan affects the trp operon.
- Compare how the presense or absence of lactose affects the lac operon.
- Compare how the presense or absence of an inducer affects activators.
- Briefly describe how small RNAs can regulate enzyme activity.
- Define the following:
- repressor
- inducer
- activator
- enhancer
- small RNAs
- Compare and contrast competitive inhibition with noncompetitive inhibition.
In living cells, there are hundreds of different enzymes working together in a coordinated manner. Living cells neither synthesize nor break down more material than is required for normal metabolism and growth. All of this necessitates precise control mechanisms for turning metabolic reactions on and off. There is tremendous diversity in the mechanisms bacteria use to regulate enzyme synthesis and enzyme activity. For pretty much every step between the activation of a gene and the final enzyme reaction from that gene product there is some bacterial mechanism for regulation that step. Here we will look at several well studied examples.
Genetic Control of Enzyme Synthesis through Repression, Induction, or Enhancement of Transcription
Genetic control of enzyme activity refers to controlling transcription of the mRNA needed for an enzyme's synthesis. In prokaryotic cells, this involves the induction, repression, or enhancement of enzyme synthesis by regulatory proteins that can bind to DNA and either induce, block, or enhance the function of RNA polymerase , the enzyme required for transcription. The regulatory proteins are often part of either an operon or a regulon. An operon is a set of genes transcribed as a polycistronic message that is collectively controlled by a regulatory protein. A regulon is a set of related genes controlled by the same regulatory protein but transcribed as monocistronic units. Regulatory proteins may function either as repressors, activators, or enhancers.
a. Repressors
Repressors are regulatory proteins that block transcription of mRNA. They do this by binding to a portion of DNA called the operator (operators are often called boxes now) that lies downstream of a promoter. The binding of the regulatory protein to the operator prevents RNA polymerase from binding to the promoter and transcribing the coding sequence for the enzymes. This is called negative control and is mostly n in biosynthetic reactions where a bacterium only makes a molecule like a particular amino acid when that amino acid is not present in the cell.
Repressors are allosteric proteins that have a binding site for a specific molecule. Binding of that molecule to the allosteric site of the repressor can alter the repressor's shape that, in turn affects its ability to bind to DNA. This can work in one of two ways:
1. Some repressors are synthesized in a form that cannot by itself bind to the operator. This is referred to as a repressible system. The binding of a molecule called a corepressor, however, alters the shape of the regulatory protein to a form that can bind to the operator and subsequently block transcription. An example of this type of repressible system is the trp operon in Escherichia coli that encodes the five enzymes in the pathway for the biosynthesis of the amino acid tryptophan. In this case, the repressor protein coded for by the trp regulatory gene, normally does not bind to the operator region of the trp operon and the five enzymes needed to synthesize the amino acid tryptophan are made (Figure \(\PageIndex{1}\)A and Figure \(\PageIndex{1}\)B).
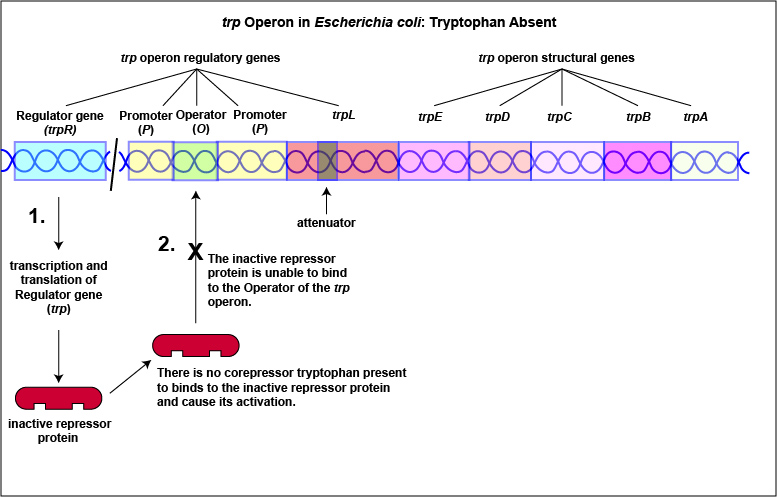
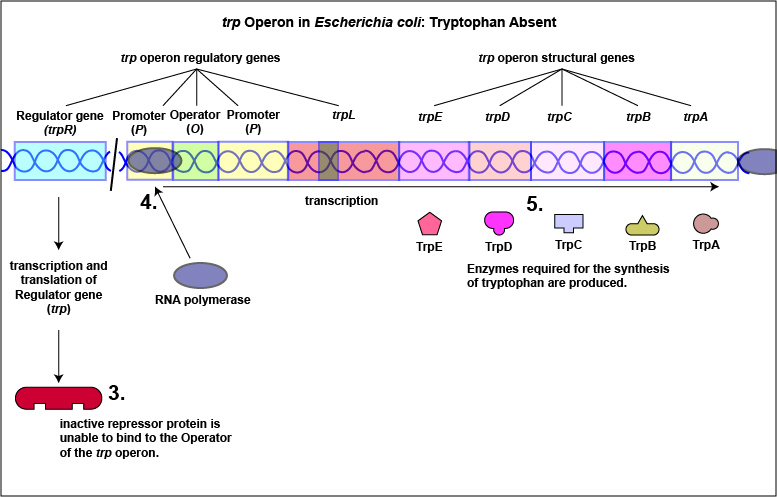
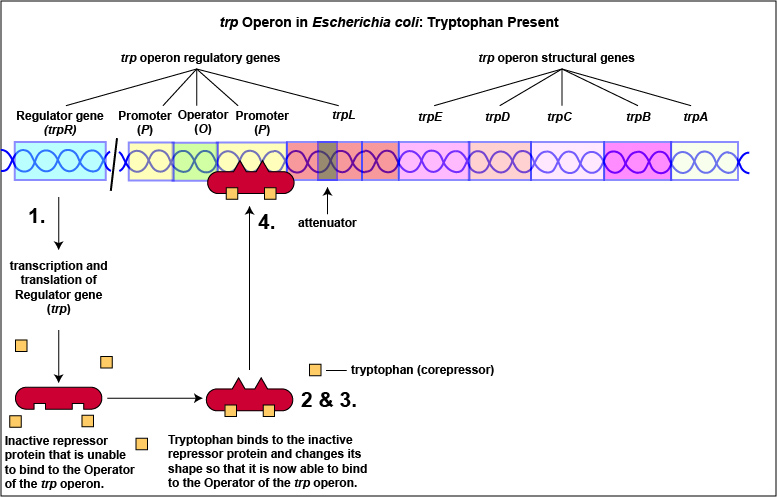
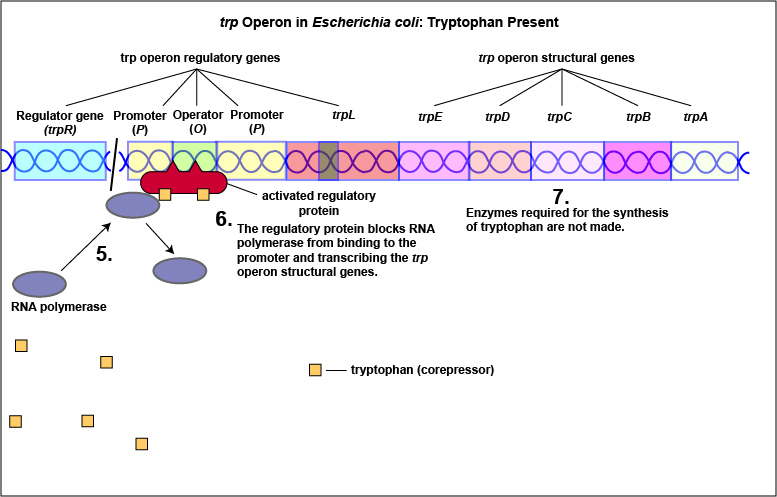
Tryptophan, the end product of these enzyme reactions, however, functions as a corepressor. Once sufficient tryptophan has been synthesized, the cell needs to terminate its synthesis. The tryptophan is able to bind to a site on the allosteric repressor protein, changing its shape and enabling it to interact with the trp operator region. Once the repressor binds to the operator, RNA polymerase is unable to bind to the promoter and transcribe the genes for tryptophan biosynthesis. Therefore, when sufficient tryptophan is present, transcription of the enzymes that allows for its biosynthesis are turned off ( Figure \(\PageIndex{2}\)A and Figure \(\PageIndex{2}\)B).
In addition to repression, the expression of the trp operon is also regulated by attenuation. The trpL gene codes for a mRNA leader sequence that controls operon expression through attenuation. This leader sequence mRNA consists of domains 1, 2, 3, and 4. Domain 3 can base pair with either domain 2 or domain 4.
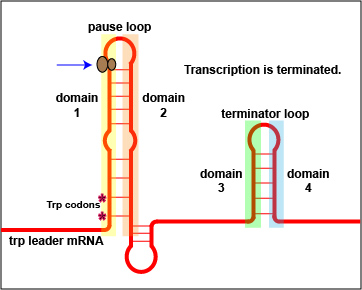
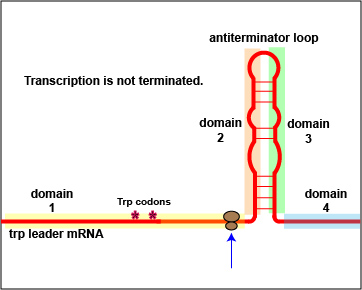
At high tryptophan concentrations, domains 3 and 4 pair in such a way as to form stem and loop structures that block the transcription of the remainder of the leader sequence mRNA and subsequently, the transcription of the structural genes for tryptophan biosynthesis ( Figure \(\PageIndex{3}\)A). However, at low concentrations of tryptophan, domains 3 and 2 pair. This pairing allows for the full transcription of the leader sequence mRNA, as well as that of the structural genes for tryptophan biosynthesis ( Figure \(\PageIndex{3}\)B).
2. Other repressors are synthesized in a form that readily binds to the operator and blocks transcription. However, the binding of a molecule called an inducer alters the shape of the regulatory protein in a way that now blocks its binding to the operator and thus permits transcription. This is referred to as an inducible system.
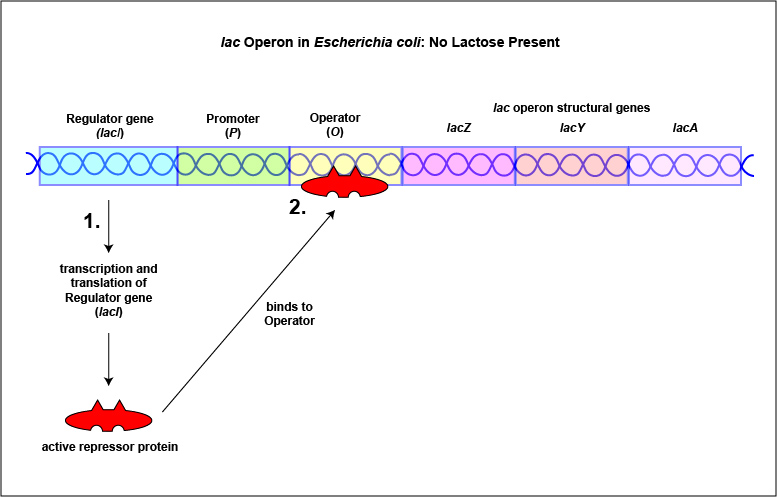
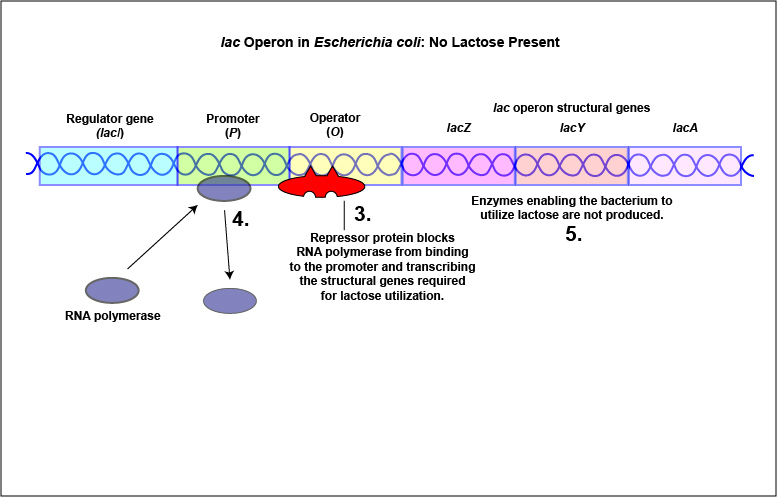
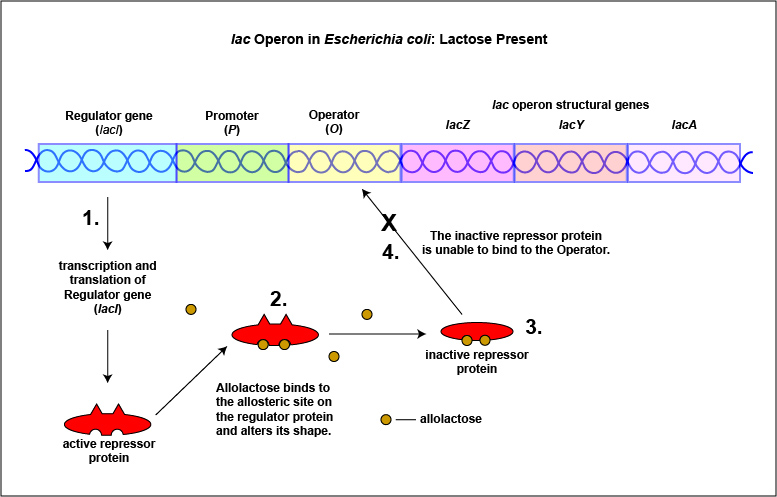
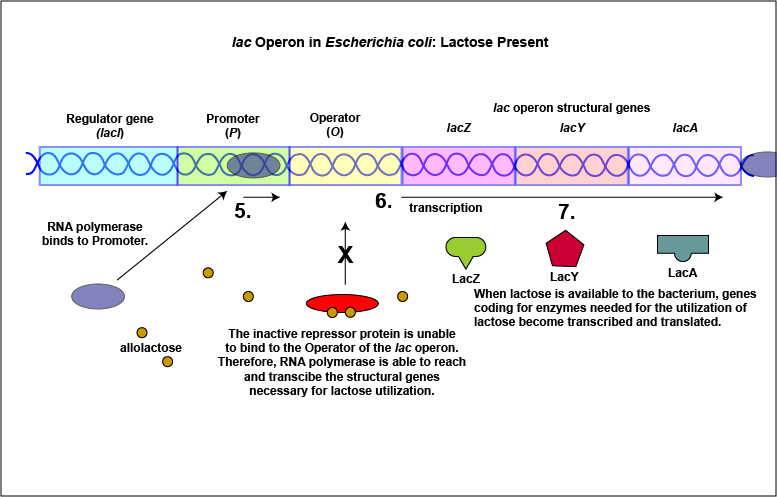
An example of an inducible system is the lac operon that encodes for the three enzymes needed for the degradation of lactose by E. coli. E. coli will only synthesize the enzymes it requires to utilize lactose if that sugar is present in the surrounding environment. In this case, lactose functions as an inducer . In the absence of lactose, the active repressor protein binds to the operator and RNA polymerase is unable to bind to the promoter and transcribe the genes for utilization of lactose. As a result, the enzymes needed for the utilization of lactose are not synthesized (Figure \(\PageIndex{4}\)A and Figure \(\PageIndex{4}\)B). When lactose, the inducer, is present, a metabolite of lactose called allolactose binds to the allosteric repressor protein and causes it to change shape in such a way that it is no longer able to bind to the operator. Now RNA polymerase is able to transcribe the three lac operon structural genes and the bacterium is able to synthesize the enzymes required for the utilization of lactose (Figure \(\PageIndex{5}\)A and Figure \(\PageIndex{5}\)B).
b. Activators
Activators are regulatory proteins that promote transcription of mRNA. Activators control genes that have a promotor to which RNA polymerase cannot bind. The promotor lies adjacent to a segment of DNA called the activator-binding site. The activator is an allosteric protein synthesized in a form that cannot normally bind to the activator-binding site. As a result, RNA polymerase is unable to bind to the promoter and transcribe the genes ( Figure \(\PageIndex{6}\)). However, binding of a molecule called an inducer to the activator alters the shape of the activator in a way that now allows it to bind to the activator-binding site. The binding of the activator to the activator-binding site, in turn, enables RNA polymerase to bind to the promotor and initiate transcription ( Figure \(\PageIndex{7}\)A and Figure \(\PageIndex{7}\)B). This is called positive control and is mostly n in catabolic reactions where a bacterium only makes enzymes for the catabolism of a substrate when that substrate is available to the cell.
c. Enhancers
Enhancers are regulatory proteins that bind to DNA located some distance from the operon they control by working with DNA-bending proteins. The DNA-binding proteins bend the DNA in a way that now allows the enhancer to interact with the promoter in such a way that RNA polymerase can now bind and initiate transcription ( Figure \(\PageIndex{8}\)).
2. Genetic Control of Enzyme Synthesis through Promoter Recognition and through DNA Supercoiling
a. Promoter Recognition: The specific sigma factors that bind to RNA polymerase determine which operon will be transcribed.
b. DNA Supercoiling: DNA supercoiling can change the tertiary shape of a DNA molecule from its normal form to one that has a left-handed twist called Z-DNA. The activities of some promoters are decreased with Z-DNA while others are increased.
3. Genetic Control of Enzyme Synthesis through the Translational Control of Enzyme Synthesis
a. RNA interference (RNAi)
RNA interference (RNAi) is a process whereby small non-coding regulatory RNAs (ncRNAs) such as microRNAs (miRNAs) regulate gene expression. These ncRNAs are regulatory molecules that are complementary to an early portion of the 5' end of the mRNA coding for the enzyme. When the small RNA binds to the mRNA by complementary base pairing , ribosomes cannot attach to the mRNA blocking its translation. As a result, the enzyme is not made ( Figure \(\PageIndex{9}\)). In bacteria these ncRNAs are often called small RNAs (sRNAs); in animal cells, plant cells, and viruses they are often called microRNAs (miRNA).
b. Ribosomal Proteins (r-proteins)
Ribosomal proteins bind to rRNA to form ribosomal subunits. Because the nucleotide base sequence for the mRNA coding for the r-proteins has similarities to that of the rRNA to which that r-protein binds during subunit formation, r-proteins not yet incorporated into ribosomal subunits can bind to that mRNA and block translation
4. Controlling the Enzyme's Activity (Feedback Inhibition).
Enzyme activity can be controlled by competitive inhibition and non-competitive inhibition.
a. With what is termed non-competitive inhibition , the inhibitor is the end product of a metabolic pathway that is able to bind to a second site (the allosteric site) on the enzyme. Binding of the inhibitor to the allosteric site alters the shape of the enzyme's active site thus preventing binding of the first substrate in the metabolic pathway. In this way, the pathway is turned off ( Figure \(\PageIndex{10}\)).
b. In the case of what is called competitive inhibition , the inhibitor is the end product of an enzymatic reaction. That end product is also capable of reacting with the enzyme's active site and prevents the enzyme from binding its normal substrate. As a result, the end product is no longer synthesized ( Figure \(\PageIndex{11}\)).
Summary
- In living cells there are hundreds of different enzymes working together in a coordinated manner, and since cells neither synthesize nor break down more material than is required for normal metabolism and growth, precise enzyme regulation is required for turning metabolic reactions on and off.
- There is tremendous diversity in the mechanisms bacteria use to regulate enzyme synthesis and enzyme activity.
- Ways in which enzymes can be controlled or regulated include controlling the synthesis of the enzyme (genetic control) and controlling the activity of the enzyme (feedback inhibition).
- In prokaryotes, genetic control of enzyme activity includes the induction or repression of enzyme synthesis by regulatory proteins that can bind to DNA and either block or enhance the function of RNA polymerase, the enzyme required for transcription.
- An operon is a set of genes collectively controlled by a regulatory protein.
- Regulatory proteins may function either as repressors or activators.
- Repressors are regulatory proteins that block transcription of mRNA by preventing RNA polymerase from transcribing the coding sequence for the enzymes.
- Some repressors, as in the case of the trp operon, are synthesized in a form that cannot by itself bind to the operator. This is referred to as a repressible system. The binding of a molecule called a corepressor, however, alters the shape of the regulatory protein to a form that can bind to the operator and subsequently block transcription.
- Some repressors, as in the case of the lac operon, are synthesized in a form that readily binds to the operator and blocks transcription. However, the binding of a molecule called an inducer alters the shape of the regulatory protein in a way that now blocks its binding to the operator and thus permits transcription. This is referred to as an inducible system.
- Activators are regulatory proteins that promote transcription of mRNA by enabling RNA polymerase to transcribing the coding sequence for the enzymes.
- Enhancers are regulatory proteins that bind to DNA located some distance from the operon they control by working with DNA-bending proteins. The DNA-bending proteins bend the DNA in a way that now allows the enhancer to interact with the promoter in such a way that RNA polymerase can now bind and initiate transcription
- Bacteria also use translational control of enzyme synthesis. One method is for the bacteria to produce noncoding RNA (ncRNA) molecules that are complementary to the mRNA coding for the enzyme, and when the small RNA binds to the mRNA by complementary base pairing, ribosomes cannot attach to the mRNA, the mRNA is not transcribed and translated into protein, and the enzyme is not made. In bacteria, these ncRNAs are often called small RNAs (sRNAs).
- Feedback inhibition controls the activity of the enzyme rather than its synthesis and can be noncompetitive or competitive.
- In the case of non-competitive inhibition, the inhibitor is the end product of a metabolic pathway that is able to bind the allosteric site on the enzyme. Binding of the inhibitor to the allosteric site alters the shape of the enzyme's active site thus preventing binding of the first substrate in the metabolic pathway. In this way, the pathway is turned off.
- In the case of what is called competitive inhibition, the inhibitor is the end product of an enzymatic reaction. That end product is also capable of reacting with the enzyme's active site and prevents the enzyme from binding its normal substrate. As a result, the end product is no longer synthesized.


