3.4: Proteins
- Page ID
- 1791
Skills to Develop
- Describe the functions proteins perform in the cell and in tissues
- Discuss the relationship between amino acids and proteins
- Explain the four levels of protein organization
- Describe the ways in which protein shape and function are linked
Proteins are one of the most abundant organic molecules in living systems and have the most diverse range of functions of all macromolecules. Proteins may be structural, regulatory, contractile, or protective; they may serve in transport, storage, or membranes; or they may be toxins or enzymes. Each cell in a living system may contain thousands of proteins, each with a unique function. Their structures, like their functions, vary greatly. They are all, however, polymers of amino acids, arranged in a linear sequence.
Types and Functions of Proteins
Enzymes, which are produced by living cells, are catalysts in biochemical reactions (like digestion) and are usually complex or conjugated proteins. Each enzyme is specific for the substrate (a reactant that binds to an enzyme) it acts on. The enzyme may help in breakdown, rearrangement, or synthesis reactions. Enzymes that break down their substrates are called catabolic enzymes, enzymes that build more complex molecules from their substrates are called anabolic enzymes, and enzymes that affect the rate of reaction are called catalytic enzymes. It should be noted that all enzymes increase the rate of reaction and, therefore, are considered to be organic catalysts. An example of an enzyme is salivary amylase, which hydrolyzes its substrate amylose, a component of starch.
Hormones are chemical-signaling molecules, usually small proteins or steroids, secreted by endocrine cells that act to control or regulate specific physiological processes, including growth, development, metabolism, and reproduction. For example, insulin is a protein hormone that helps to regulate the blood glucose level. The primary types and functions of proteins are listed in Table \(\PageIndex{1}\).
| Type | Examples | Functions |
|---|---|---|
| Digestive Enzymes | Amylase, lipase, pepsin, trypsin | Help in digestion of food by catabolizing nutrients into monomeric units |
| Transport | Hemoglobin, albumin | Carry substances in the blood or lymph throughout the body |
| Structural | Actin, tubulin, keratin | Construct different structures, like the cytoskeleton |
| Hormones | Insulin, thyroxine | Coordinate the activity of different body systems |
| Defense | Immunoglobulins | Protect the body from foreign pathogens |
| Contractile | Actin, myosin | Effect muscle contraction |
| Storage | Legume storage proteins, egg white (albumin) | Provide nourishment in early development of the embryo and the seedling |
Proteins have different shapes and molecular weights; some proteins are globular in shape whereas others are fibrous in nature. For example, hemoglobin is a globular protein, but collagen, found in our skin, is a fibrous protein. Protein shape is critical to its function, and this shape is maintained by many different types of chemical bonds. Changes in temperature, pH, and exposure to chemicals may lead to permanent changes in the shape of the protein, leading to loss of function, known as denaturation. All proteins are made up of different arrangements of the same 20 types of amino acids.
Amino Acids
Amino acids are the monomers that make up proteins. Each amino acid has the same fundamental structure, which consists of a central carbon atom, also known as the alpha (α) carbon, bonded to an amino group (NH2), a carboxyl group (COOH), and to a hydrogen atom. Every amino acid also has another atom or group of atoms bonded to the central atom known as the R group (Figure \(\PageIndex{1}\)).
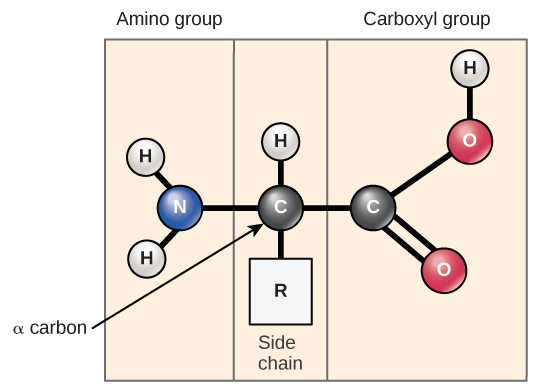
The name "amino acid" is derived from the fact that they contain both amino group and carboxyl-acid-group in their basic structure. As mentioned, there are 20 amino acids present in proteins. Ten of these are considered essential amino acids in humans because the human body cannot produce them and they are obtained from the diet. For each amino acid, the R group (or side chain) is different (Figure \(\PageIndex{2}\)).
Art Connection
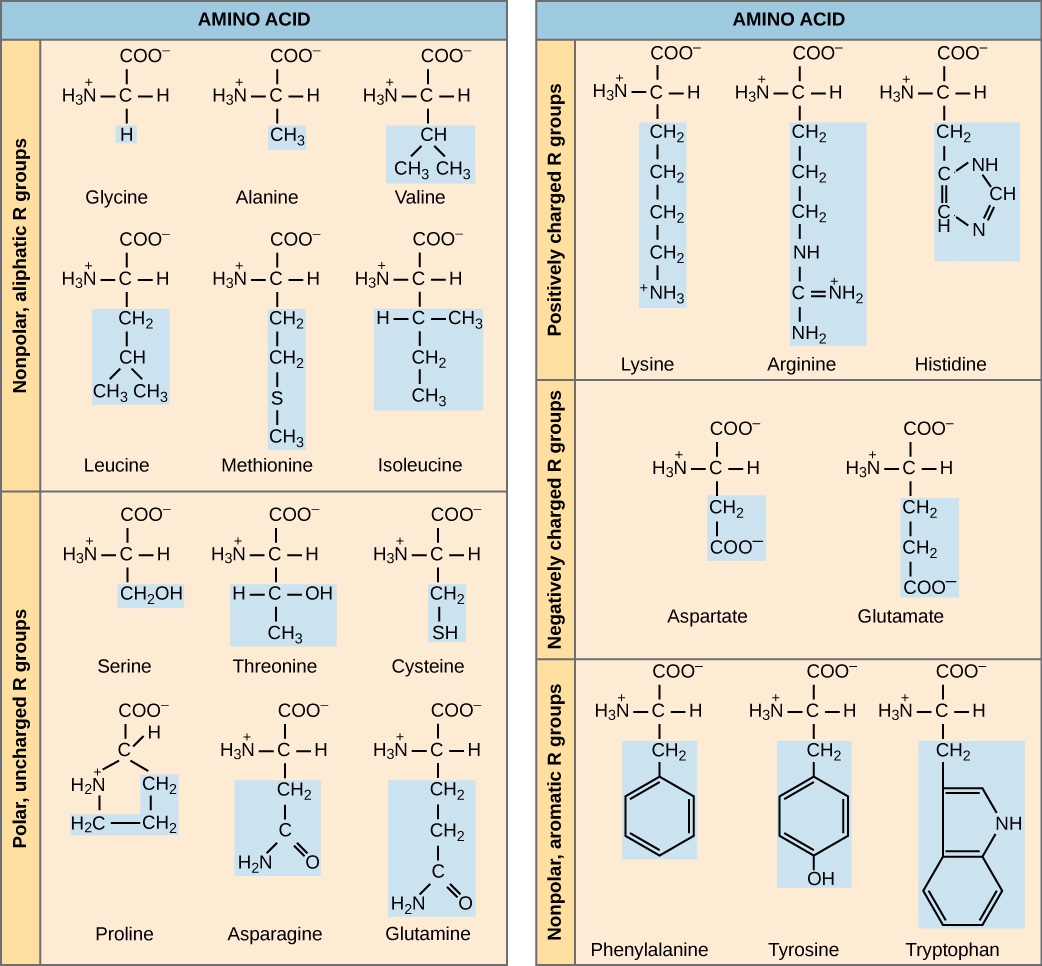
Which categories of amino acid would you expect to find on the surface of a soluble protein, and which would you expect to find in the interior? What distribution of amino acids would you expect to find in a protein embedded in a lipid bilayer?
The chemical nature of the side chain determines the nature of the amino acid (that is, whether it is acidic, basic, polar, or nonpolar). For example, the amino acid glycine has a hydrogen atom as the R group. Amino acids such as valine, methionine, and alanine are nonpolar or hydrophobic in nature, while amino acids such as serine, threonine, and cysteine are polar and have hydrophilic side chains. The side chains of lysine and arginine are positively charged, and therefore these amino acids are also known as basic amino acids. Proline has an R group that is linked to the amino group, forming a ring-like structure. Proline is an exception to the standard structure of an animo acid since its amino group is not separate from the side chain (Figure \(\PageIndex{2}\)).
Amino acids are represented by a single upper case letter or a three-letter abbreviation. For example, valine is known by the letter V or the three-letter symbol val. Just as some fatty acids are essential to a diet, some amino acids are necessary as well. They are known as essential amino acids, and in humans they include isoleucine, leucine, and cysteine. Essential amino acids refer to those necessary for construction of proteins in the body, although not produced by the body; which amino acids are essential varies from organism to organism.
The sequence and the number of amino acids ultimately determine the protein's shape, size, and function. Each amino acid is attached to another amino acid by a covalent bond, known as a peptide bond, which is formed by a dehydration reaction. The carboxyl group of one amino acid and the amino group of the incoming amino acid combine, releasing a molecule of water. The resulting bond is the peptide bond (Figure \(\PageIndex{3}\)).
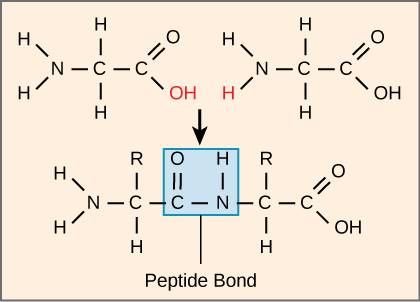
The products formed by such linkages are called peptides. As more amino acids join to this growing chain, the resulting chain is known as a polypeptide. Each polypeptide has a free amino group at one end. This end is called the N terminal, or the amino terminal, and the other end has a free carboxyl group, also known as the C or carboxyl terminal. While the terms polypeptide and protein are sometimes used interchangeably, a polypeptide is technically a polymer of amino acids, whereas the term protein is used for a polypeptide or polypeptides that have combined together, often have bound non-peptide prosthetic groups, have a distinct shape, and have a unique function. After protein synthesis (translation), most proteins are modified. These are known as post-translational modifications. They may undergo cleavage, phosphorylation, or may require the addition of other chemical groups. Only after these modifications is the protein completely functional.
Evolution Connection
The Evolutionary Significance of Cytochrome cCytochrome c is an important component of the electron transport chain, a part of cellular respiration, and it is normally found in the cellular organelle, the mitochondrion. This protein has a heme prosthetic group, and the central ion of the heme gets alternately reduced and oxidized during electron transfer. Because this essential protein’s role in producing cellular energy is crucial, it has changed very little over millions of years. Protein sequencing has shown that there is a considerable amount of cytochrome c amino acid sequence homology among different species; in other words, evolutionary kinship can be assessed by measuring the similarities or differences among various species’ DNA or protein sequences.
Scientists have determined that human cytochrome c contains 104 amino acids. For each cytochrome c molecule from different organisms that has been sequenced to date, 37 of these amino acids appear in the same position in all samples of cytochrome c. This indicates that there may have been a common ancestor. On comparing the human and chimpanzee protein sequences, no sequence difference was found. When human and rhesus monkey sequences were compared, the single difference found was in one amino acid. In another comparison, human to yeast sequencing shows a difference in the 44th position.
Protein Structure
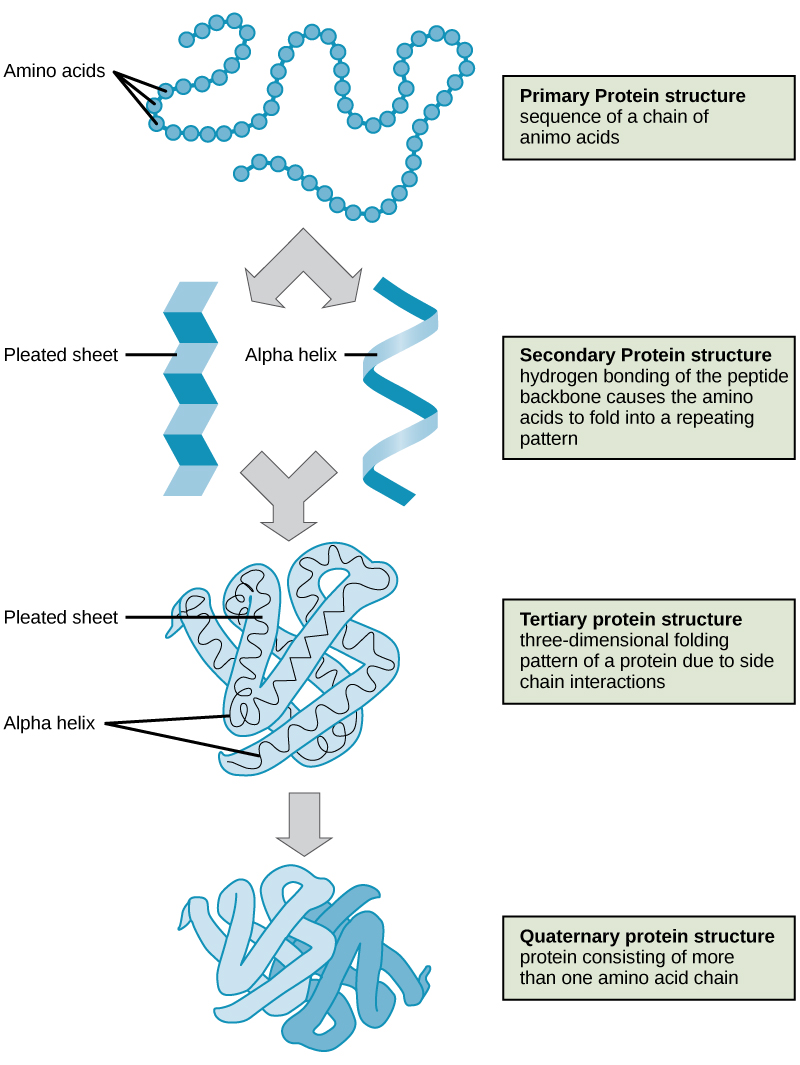
As discussed earlier, the shape of a protein is critical to its function. For example, an enzyme can bind to a specific substrate at a site known as the active site. If this active site is altered because of local changes or changes in overall protein structure, the enzyme may be unable to bind to the substrate. To understand how the protein gets its final shape or conformation, we need to understand the four levels of protein structure: primary, secondary, tertiary, and quaternary.
Primary Structure
The unique sequence of amino acids in a polypeptide chain is its primary structure. For example, the pancreatic hormone insulin has two polypeptide chains, A and B, and they are linked together by disulfide bonds. The N terminal amino acid of the A chain is glycine, whereas the C terminal amino acid is asparagine (Figure \(\PageIndex{4}\)). The sequences of amino acids in the A and B chains are unique to insulin.

The unique sequence for every protein is ultimately determined by the gene encoding the protein. A change in nucleotide sequence of the gene’s coding region may lead to a different amino acid being added to the growing polypeptide chain, causing a change in protein structure and function. In sickle cell anemia, the hemoglobin β chain (a small portion of which is shown in Figure \(\PageIndex{5}\)) has a single amino acid substitution, causing a change in protein structure and function. Specifically, the amino acid glutamic acid is substituted by valine in the β chain. What is most remarkable to consider is that a hemoglobin molecule is made up of two alpha chains and two beta chains that each consist of about 150 amino acids. The molecule, therefore, has about 600 amino acids. The structural difference between a normal hemoglobin molecule and a sickle cell molecule—which dramatically decreases life expectancy—is a single amino acid of the 600. What is even more remarkable is that those 600 amino acids are encoded by three nucleotides each, and the mutation is caused by a single base change (point mutation), 1 in 1800 bases.

Because of this change of one amino acid in the chain, hemoglobin molecules form long fibers that distort the biconcave, or disc-shaped, red blood cells and assume a crescent or “sickle” shape, which clogs arteries (Figure \(\PageIndex{6}\)). This can lead to myriad serious health problems such as breathlessness, dizziness, headaches, and abdominal pain for those affected by this disease.

Secondary Structure
The local folding of the polypeptide in some regions gives rise to the secondary structure of the protein. The most common are the α-helix and β-pleated sheet structures (Figure \(\PageIndex{7}\)). Both structures are the α-helix structure—the helix held in shape by hydrogen bonds. The hydrogen bonds form between the oxygen atom in the carbonyl group in one amino acid and another amino acid that is four amino acids farther along the chain.

Every helical turn in an alpha helix has 3.6 amino acid residues. The R groups (the variant groups) of the polypeptide protrude out from the α-helix chain. In the β-pleated sheet, the “pleats” are formed by hydrogen bonding between atoms on the backbone of the polypeptide chain. The R groups are attached to the carbons and extend above and below the folds of the pleat. The pleated segments align parallel or antiparallel to each other, and hydrogen bonds form between the partially positive nitrogen atom in the amino group and the partially negative oxygen atom in the carbonyl group of the peptide backbone. The α-helix and β-pleated sheet structures are found in most globular and fibrous proteins and they play an important structural role.
Tertiary Structure
The unique three-dimensional structure of a polypeptide is its tertiary structure (Figure \(\PageIndex{8}\)). This structure is in part due to chemical interactions at work on the polypeptide chain. Primarily, the interactions among R groups creates the complex three-dimensional tertiary structure of a protein. The nature of the R groups found in the amino acids involved can counteract the formation of the hydrogen bonds described for standard secondary structures. For example, R groups with like charges are repelled by each other and those with unlike charges are attracted to each other (ionic bonds). When protein folding takes place, the hydrophobic R groups of nonpolar amino acids lay in the interior of the protein, whereas the hydrophilic R groups lay on the outside. The former types of interactions are also known as hydrophobic interactions. Interaction between cysteine side chains forms disulfide linkages in the presence of oxygen, the only covalent bond forming during protein folding.
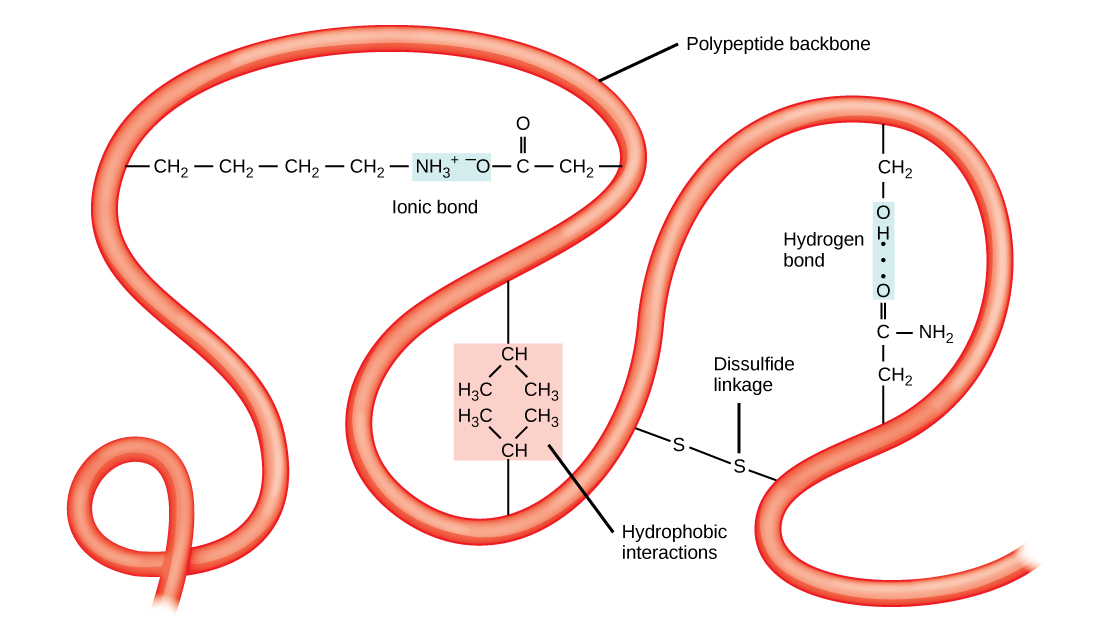
All of these interactions, weak and strong, determine the final three-dimensional shape of the protein. When a protein loses its three-dimensional shape, it may no longer be functional.
Quaternary Structure
In nature, some proteins are formed from several polypeptides, also known as subunits, and the interaction of these subunits forms the quaternary structure. Weak interactions between the subunits help to stabilize the overall structure. For example, insulin (a globular protein) has a combination of hydrogen bonds and disulfide bonds that cause it to be mostly clumped into a ball shape. Insulin starts out as a single polypeptide and loses some internal sequences in the presence of post-translational modification after the formation of the disulfide linkages that hold the remaining chains together. Silk (a fibrous protein), however, has a β-pleated sheet structure that is the result of hydrogen bonding between different chains.
The four levels of protein structure (primary, secondary, tertiary, and quaternary) are illustrated in Figure \(\PageIndex{9}\).
Denaturation and Protein Folding
Each protein has its own unique sequence and shape that are held together by chemical interactions. If the protein is subject to changes in temperature, pH, or exposure to chemicals, the protein structure may change, losing its shape without losing its primary sequence in what is known as denaturation. Denaturation is often reversible because the primary structure of the polypeptide is conserved in the process if the denaturing agent is removed, allowing the protein to resume its function. Sometimes denaturation is irreversible, leading to loss of function. One example of irreversible protein denaturation is when an egg is fried. The albumin protein in the liquid egg white is denatured when placed in a hot pan. Not all proteins are denatured at high temperatures; for instance, bacteria that survive in hot springs have proteins that function at temperatures close to boiling. The stomach is also very acidic, has a low pH, and denatures proteins as part of the digestion process; however, the digestive enzymes of the stomach retain their activity under these conditions.
Protein folding is critical to its function. It was originally thought that the proteins themselves were responsible for the folding process. Only recently was it found that often they receive assistance in the folding process from protein helpers known as chaperones (or chaperonins) that associate with the target protein during the folding process. They act by preventing aggregation of polypeptides that make up the complete protein structure, and they disassociate from the protein once the target protein is folded.
Summary
Proteins are a class of macromolecules that perform a diverse range of functions for the cell. They help in metabolism by providing structural support and by acting as enzymes, carriers, or hormones. The building blocks of proteins (monomers) are amino acids. Each amino acid has a central carbon that is linked to an amino group, a carboxyl group, a hydrogen atom, and an R group or side chain. There are 20 commonly occurring amino acids, each of which differs in the R group. Each amino acid is linked to its neighbors by a peptide bond. A long chain of amino acids is known as a polypeptide.
Proteins are organized at four levels: primary, secondary, tertiary, and (optional) quaternary. The primary structure is the unique sequence of amino acids. The local folding of the polypeptide to form structures such as the α helix and β-pleated sheet constitutes the secondary structure. The overall three-dimensional structure is the tertiary structure. When two or more polypeptides combine to form the complete protein structure, the configuration is known as the quaternary structure of a protein. Protein shape and function are intricately linked; any change in shape caused by changes in temperature or pH may lead to protein denaturation and a loss in function.
Art Connections
Figure \(\PageIndex{2}\): Which categories of amino acid would you expect to find on the surface of a soluble protein, and which would you expect to find in the interior? What distribution of amino acids would you expect to find in a protein embedded in a lipid bilayer?
- Answer
-
Polar and charged amino acid residues (the remainder after peptide bond formation) are more likely to be found on the surface of soluble proteins where they can interact with water, and nonpolar (e.g., amino acid side chains) are more likely to be found in the interior where they are sequestered from water. In membrane proteins, nonpolar and hydrophobic amino acid side chains associate with the hydrophobic tails of phospholipids, while polar and charged amino acid side chains interact with the polar head groups or with the aqueous solution. However, there are exceptions. Sometimes, positively and negatively charged amino acid side chains interact with one another in the interior of a protein, and polar or charged amino acid side chains that interact with a ligand can be found in the ligand binding pocket.
Glossary
- alpha-helix structure (α-helix)
- type of secondary structure of proteins formed by folding of the polypeptide into a helix shape with hydrogen bonds stabilizing the structure
- amino acid
- monomer of a protein; has a central carbon or alpha carbon to which an amino group, a carboxyl group, a hydrogen, and an R group or side chain is attached; the R group is different for all 20 amino acids
- beta-pleated sheet (β-pleated)
- secondary structure found in proteins in which “pleats” are formed by hydrogen bonding between atoms on the backbone of the polypeptide chain
- chaperone
- (also, chaperonin) protein that helps nascent protein in the folding process
- denaturation
- loss of shape in a protein as a result of changes in temperature, pH, or exposure to chemicals
- enzyme
- catalyst in a biochemical reaction that is usually a complex or conjugated protein
- hormone
- chemical signaling molecule, usually protein or steroid, secreted by endocrine cells that act to control or regulate specific physiological processes
- peptide bond
- bond formed between two amino acids by a dehydration reaction
- polypeptide
- long chain of amino acids linked by peptide bonds
- primary structure
- linear sequence of amino acids in a protein
- protein
- biological macromolecule composed of one or more chains of amino acids
- quaternary structure
- association of discrete polypeptide subunits in a protein
- secondary structure
- regular structure formed by proteins by intramolecular hydrogen bonding between the oxygen atom of one amino acid residue and the hydrogen attached to the nitrogen atom of another amino acid residue
- tertiary structure
- three-dimensional conformation of a protein, including interactions between secondary structural elements; formed from interactions between amino acid side chains


