18.3: The Evolution of Body Form in Animals
- Page ID
- 5920
Advances in genetics - especially the sequencing of entire genomes from a wide variety of animals - has revealed an unexpected paradox. While the animal kingdom contains an extraordinary diversity of body types:
- with sizes ranging from the microscopic daphnia (a crustacean) to the great blue whale
- with body plans as diverse as those of Drosophila and humans,
the great structural diversity of animals is not reflected in their genetic makeup. Throughout the animal kingdom, one finds thousands of orthologous genes; that is, genes that have similar sequences and encode similar products.
At the level of the cell, and even of tissues, this perhaps should not be surprising. After all, most types of cells - from whatever animal - are quite similar in their structure and function. Thus we would expect that their genes that encode ribosomal proteins, cytochromes, histones, etc., etc. would be similar.
In trying to resolve the paradox that these findings present, it is useful to distinguish between
- "housekeeping" genes - genes that encode proteins (e.g. cytochromes) and RNAs (e.g. ribosomal RNAs) that function in all cells and
- "toolkit" genes
- genes that control the expression of other genes by encoding transcription factors
- genes that encode cell-signaling proteins that signal the cell to turn on (or off) a genetic program.
Housekeeping genes generally
- show modest sequence differences from animal to animal. Some of these are neutral in their effect on the gene's function while others are the result of evolutionary adaptation. Both can be used to trace taxonomic relationships.
- have simple control regions, i.e. promoters and enhancers.
- are expressed in all types of cells in the body (almost half of our protein-encoding genes are expressed in every cell - liver, neurons, muscle, etc.).
Toolkit genes generally
- show very slight sequence differences between different animal species in their coding regions (exons) while
- they have very elaborate control regions with many promoter and enhancer sites each of which has a binding site for one or another different transcription factors.
The proteins (transcription factors and cell-signaling molecules) of toolkit genes are identical in whatever cells they are expressed in. However, the function of that protein can vary greatly
- in different tissues of the animal - a phenomenon called mosaic pleiotropy
- at different times in the embryonic development of the animal - a phenomenon called heterochrony
Even such primitive animals as sponges and cnidarians have hundreds of toolkit genes that are clear orthologs to genes of humans. Some examples are genes whose products are involved in cell signaling (e.g., Wnt and β-catenin, Hedgehog, Notch, Receptor Tyrosine Kinases (RTKs), components of JAK/STAT pathways, and Transforming Growth Factor-beta TGF-β receptors).
In fact the sequences of many of these genes from different animal phyla are so similar that they can be interchanged. This similarity can be tested in animals that can be made transgenic. Some examples:
- The mouse gene Pax6 (also known as small eyes [Sey] for the mutant phenotype) can substitute for the mutant eyeless gene in Drosophila while the human PAX6 gene can restore normal eye development in the mutant small eyes (Sey) mouse.
- The mouse homeobox gene HoxB6 can substitute for the Drosophila homeobox gene Antennapedia (Antp) and when introduced into Drosophila give rise to legs in place of antennae just as mutant Antp genes do.
Mosaic Pleiotropy
Pleiotropy is the production by a single gene of more than one effect on the phenotype. Mosaic refers to the patchy distribution of cells and tissues expressing that phenotype.
An example:
Pitx1 is a
- Homeobox gene (similar to bicoid in Drosophila) with orthologs found in all vertebrates.
- It contains 3 exons that encode a protein of some 283 amino acids (varying slightly in different species) which is
- A transcription factor that regulates the expression of other genes involved in the differentiation and function of
- the anterior lobe of the pituitary gland (Pitx1 = "Pituitary homeobox1");
- jaw development (mutations are associated with cleft palate in mammals);
- development of the thymus and some types of mechanoreceptors;
- development of the hind limbs.
- Its activity in these regions is controlled by regulatory regions (promoters and/or enhancers) specific to each region (and presumably turned on by other transcription factors in the cells of those regions).
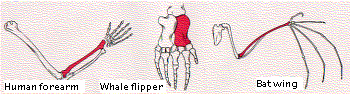
When we consider the dramatically-different activities that a given toolkit gene product can perform in different parts of the same animal, it is easier to understand how easy it must be for these same genes to alter the structure of the same body part in different species, e.g., the human arm and the wing of the bat.
Mutations in Regulatory Regions
Not all genes need to be expressed in all cells. In which cells and when a given gene will be expressed is controlled by the interaction of:
- extracellular signals turning on (or off)
- transcription factors, which turn on (or off)
- particular genes
A mutation that would be lethal in the protein coding region of a gene need not be if it occurs in a control region (e.g. promoters and/or enhancers) of that gene. In fact, there is increasing evidence that mutations in control regions have played an important part in evolution. Examples:
- Humans have a gene (LCT) encoding lactase; the enzyme that digests lactose (e.g. in milk). In most of the world's people, LCT is active in young children but is turned off in adults. However, northern Europeans and three different tribes of African pastoralists, for whom milk remains a part of the adult diet, carry a mutation in the control region of their lactase gene that permits it to be expressed in adults. The mutation is different in each of the 4 cases — examples of convergent evolution.
- There are very few differences in the coding sequences between genes of humans and chimpanzees. However, many of their shared genes differ in their control regions.
- The story of Prx1. Prx1 encodes a transcription factor that is essential for forelimb growth in mammals. When mice have the enhancer region of their Prx1 replaced with the enhancer region of Prx1 from a bat (whose front limbs are wings), the front legs of resulting mice are 6% longer than normal. Here, then is a morphological change not driven by a change in the Prx1 protein but by a change in the expression of its gene.
- The story of Pitx1.
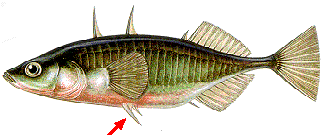
In a remarkable study of three-spined sticklebacks published in the 15 April 2004 issue of Nature, Michael Shapiro, Melissa Marks, Catherine Peichel, and their colleagues report that a mutation in a noncoding region of the Pitx1 gene accounts for most of the difference in the structure of the pelvic bones of the marine stickleback and its close freshwater cousins.
The marine sticklebacks
- have prominent spines jutting out in their pelvic region (red arrow) as well as the spines along the back (that give the fish its name). These spines may help protect them from being eaten by predators. (Drawing courtesy of the Parks Administration in the Emilia-Romagna region of Italy.)
- express the Pitx1 gene in various tissues, including
- thymus
- mechanoreceptors
- the pelvic region
The freshwater sticklebacks
- have no or very much smaller spines in their pelvic region
- express the identical Pitx1 gene in all the same tissues except those that develop into the pelvic structures
- The reason: a deletion in an enhancer of Pitx1 responsible for turning on Pitx1 in the developing pelvic area. (Mice homozygous for a mutation in this control region have deformed hind limbs.)
Here then is a remarkable demonstration of how a single gene mutation can not only be viable but can lead to a major change in phenotype - adaptive evolution. (The changes seem not to have produce true speciation as yet. The marine and freshwater forms can interbreed. In fact, that is how the differences in their hind limbs were found to be primarily due to the expression of Pitx1.)
Heterochrony
What toolkit proteins do is governed not only by what tissue they are being produced in but also by when they are produced - a phenomenon called heterochrony.
Examples:
- The transcription factor decapentaplegic (DPP) plays a wide variety of roles in the development of Drosophila from laying the foundation for the future central nervous system in the early embryo to the elaboration of wings, legs, antennae, etc. in the adult.
- The formation of vertebrae. As their name implies, a key feature of all vertebrates is their backbone of vertebrae. However, the number of these can vary greatly. Humans have 33 while snakes can have several hundred. However, the toolkit genes (e.g., Wnt, Notch, FGF-β) responsible for forming vertebrae appear to be the same for all. What makes the difference is the timing (rate) at which pulses of these proteins are produced relative to the rate at which the embryo grows.
The Tc1 Mouse
So the evidence is increasing that what makes the difference between a human and a chimpanzee (or any other pair of animals) is in large measure
- not a matter of their inheritance of different genes and their encoded proteins and RNAs but
- their inheritance of mutations in the control regions - promoters and enhancers - that regulate where and when these genes will be expressed.
A vivid example of this is the work reported by Wilson et al. in the 17 October 2008 issue of Science. Their experimental material was liver cells (hepatocytes) taken from
- normal humans
- normal mice
- the Tc1 mouse
The Tc1 mouse is more than simply transgenic, it carries in most of its cells a human chromosome #21. This small chromosome is the one that, when present in a triple dose (trisomy 21), produces Down syndrome in humans. Mice have a similar chromosome that is designated #16.
The question that this remarkable animal could answer: will the genes on human chromosome #21 (105 of them) when present in a mouse nucleus and surrounded by mouse transcription factors and signaling pathways respond as the mouse #16 does or as #21 does in human liver cells or something quite different from either?
The answer turned out that the #21 responded pretty much as it does in its normal human cellular environment.
One line of evidence
Several transcription factors turn on gene activity in liver cells. As seems to be the case with all transcription factors, the human and mouse versions are close orthologs (95% identical in sequence). Using ChIP analysis, they found that the mouse transcription factors bound to sites along the human chromosome much as the human transcription factors do. (Chromosome #21 does not encode any transcription factors, so all those available in the mouse nucleus were of mouse origin.)
| Tissue | Chromosome | Transcription Factors (TFs) | Sites Bound by TFs | Gene Expression |
|---|---|---|---|---|
| human liver cells | #21 | human TFs | human pattern | human pattern |
| Tc1 liver cells | #21 | mouse TFs | human pattern | human pattern |
| #16 | mouse TFs | mouse pattern | mouse pattern | |
| normal mouse liver cells | #16 | mouse TFs | mouse pattern | mouse pattern |
A second line of evidence: gene expression
These workers also examined the pattern of gene expression; that is, the production of messenger RNAs, in the various combinations. They did this using microarrays. The human genes expressed on chromosome #21 by mouse transcription factors in the Tc1 mouse cells were mostly the same as those turned on by human transcription factors in human cells.
The Bottom Line
All these lines of evidence point to the following:
Throughout the animal kingdom,
- Genes encoding "housekeeping" proteins (histones, enzymes, etc., etc.) are remarkably conserved; that is, their products vary only slightly - certainly not enough to account for the vast range of bodies seen in animals from sea anemones to humans.
- Genes encoding "toolkit" proteins (transcription factors, cell signaling molecules) are also highly conserved (even more so).
- So the key to the fabulous diversity in the animal kingdom must lie in what evolution has done to the DNA sequences that control where and when genes are expressed in the developing animal.
So after years of looking for and sequencing open reading frames, the task now will be to analyze the sequence differences - arisen by mutation and evolution - in the intergenic regions that serve as control regions of those genes. An early result of genome analysis: during the radiation of the various mammalian orders, enhancers have diversified (evolved) much more rapidly than have promoters and their associated protein-encoding genes.


