9.3: Regulation of Gene Expression in Eukaryotes
- Page ID
- 4878
The latest estimates are that a human cell, a eukaryotic cell, contains some 21,000 genes. Some of these are expressed in all cells all the time. These so-called housekeeping genes are responsible for the routine metabolic functions (e.g. respiration) common to all cells. Some are expressed as a cell enters a particular pathway of differentiation. Some are expressed all the time in only those cells that have differentiated in a particular way. For example, a plasma cell expresses continuously the genes for the antibody it synthesizes. Some are expressed only as conditions around and in the cell change. For example, the arrival of a hormone may turn on (or off) certain genes in that cell.
How is gene expression regulated? There are several methods used by eukaryotes.
- Altering the rate of transcription of the gene. This is the most important and widely-used strategy.
- However, eukaryotes supplement transcriptional regulation with several other methods:
- Altering the rate at which RNA transcripts are processed while still within the nucleus.
- Altering the stability of messenger RNA (mRNA) molecules; that is, the rate at which they are degraded.
- Altering the efficiency with which ribosomes translate the mRNA into a polypeptide.
Protein-coding genes have
- exons whose sequence encodes the polypeptide
- introns that will be removed from the mRNA before it is translated
- a transcription start site
- promoters
- a basal or core promoter located within about 40 base pairs (bp) of the start site
- "upstream" promoters, which may extend over as many as 200 bp farther upstream
- enhancers
- silencers
Adjacent genes are often separated by an insulator which helps them avoid cross-talk between each other's promoters and enhancers (and/or silencers).
Transcription start site
This is where a molecule of RNA polymerase II (pol II, also known as RNAP II) binds. Pol II is a complex of 12 different proteins (shown in the figure in yellow with small colored circles superimposed on it). The start site is where transcription of the gene into RNA begins.
The core promoter
All eukaryotic genes contain a core promoter. One common example is a sequence of bases (e.g., TATAAAAAA) called the TATA box. It is bound by a large complex of some 50 different proteins, including
- Transcription Factor IID (TFIID) which is a complex of
- TATA-binding protein (TBP), which recognizes and binds to the TATA box
- 13 other protein factors which bind to TBP, each other, and (some of them) to the DNA.
- Transcription Factor IIB (TFIIB) which binds both the DNA and pol II.
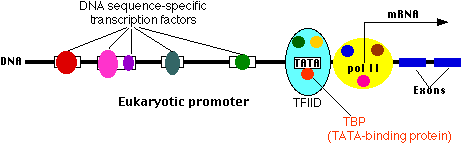
Many different genes and many different types of cells share the same transcription factors — not only those that bind at the core promoter but even some of those that bind upstream. What turns on a particular gene in a particular cell is probably the unique combination of promoter sites and the transcription factors that are chosen.
An Analogy
The rows of lock boxes in a bank provide a useful analogy. To open any particular box in the room requires two keys:
- Your key, whose pattern of notches fits only the lock of the box assigned to you (= the upstream promoter), but which cannot unlock the box without
- A key carried by a bank employee that can activate the unlocking mechanism of any box (= the core promoter) but cannot by itself open any box.
Hormones Effect
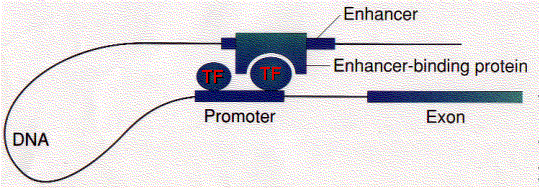
These loops are stabilized by a protein designated CTCF ("CCCTC binding factor"; named for the nucleotide sequence to which it binds). The CTCF at one site on the DNA forms a dimer with the CTCF at another site on the DNA binding the two regions together. CTCF has 11 zinc fingers. They can also be stabilized by cohesin — the same protein complex that holds sister chromatids together during mitosis and meiosis.
Michael R. Botchan and his colleagues have produced visual evidence of this model of enhancer action. They created an artificial DNA molecule with
- several (4) promoter sites for Sp1 about 300 bases from one end. Sp1 is a zinc-finger transcription factor that binds to the sequence 5' GGGCGG 3' found in the promoters of many genes, especially "housekeeping" genes.
- several (5) enhancer sites about 800 bases from the other end. These are bound by an enhancer-binding protein designated E2.
- 1860 base pairs of DNA between the two.
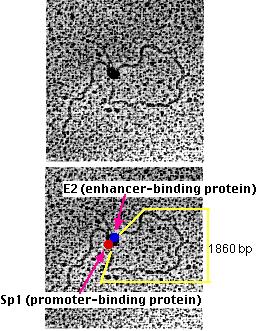
Significance of "Looping"
The looping of chromosomes that brings enhancers close to promoters (and promoters close to other promoters) seems to be a mechanism to ensure the expression (or inhibition) of groups of genes that must perform together. The response of a cell to the arrival of a signal (e.g., a hormone) may involve turning on (or off) hundreds of different genes whose products must be produced in a coordinated way for the cell to respond appropriately. The dynamic movement of portions of the chromosome carrying the appropriate gene loci into a "transcription factory" may be a mechanism to accomplish this. If so, we are seeing the eukaryotic equivalent of the coordinated gene expression provided by operons in bacteria.
Silencers
Silencers are control regions of DNA that, like enhancers, may be located thousands of base pairs away from the gene they control. However, when transcription factors bind to them, expression of the gene they control is repressed.
Insulators
As you can see above, enhancers can turn on promoters of genes located thousands of base pairs away. Insulators prevent an enhancer from inappropriately binding to and activating the promoter of some other gene in the same region of the chromosome..
Insulators are
- stretches of DNA (as few as 42 base pairs may do the trick)
- located between the
- enhancer(s) and promoter(s) or
- silencer(s) and promoter(s)
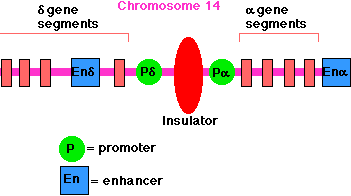
The enhancer for the promoter of the gene for the delta chain of the gamma/delta T-cell receptor for antigen (TCR) is located close to the promoter for the alpha chain of the alpha/beta TCR (on chromosome 14 in humans). A T cell must choose between one or the other. There is an insulator between the alpha gene promoter and the delta gene promoter that ensures that activation of one does not spread over to the other.
All insulators discovered so far in vertebrates work only when bound by the CTCF protein. Another example: In mammals (mice, humans, pigs), only the allele for insulin-like growth factor-2 (IGF2) inherited from one's father is active; that inherited from the mother is not — a phenomenon called imprinting.
The mechanism: the mother's allele has an insulator between the IGF2 promoter and enhancer. So does the father's allele, but in his case, the insulator has been methylated. CTCF can no longer bind to the insulator, and so the enhancer is now free to turn on the father's IGF2 promoter.
Many of the commercially-important varieties of pigs have been bred to contain a gene that increases the ratio of skeletal muscle to fat. This gene has been sequenced and turns out to be an allele of IGF2, which contains a single point mutation in one of its introns. Pigs with this mutation produce higher levels of IGF2 mRNA in their skeletal muscles (but not in their liver). This tells us that:
- Mutations need not be in the protein-coding portion of a gene in order to affect the phenotype.
- Mutations in non-coding portions of a gene can affect how that gene is regulated (here, a change in muscle but not in liver).


