9.10: Palindromes
- Page ID
- 4892
A palindrome is a sequence of letters and/or words, that reads the same forwards and backwards. "able was I ere I saw elba" is a palindrome. Palindromes also occur in a DNA. There are two types.
Palindromes that occur on opposite strands of the same section of DNA helix
5' GGCC 3'
3' CCGG 5'
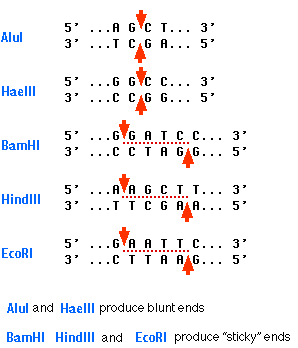
This type of palindrome serves as the target for most restriction enzymes. The graphic shows the palindromic sequences "seen" by five restriction enzymes (named in blue) commonly used in recombinant DNA work.
Inverted Repeats
In these cases, two different segments of the double helix read the same but in opposite directions.
5' AGAACAnnnTGTTCT 3'
3' TCTTGTnnnACAAGA 5'
Inverted repeats are commonly found in
- The DNA to which transcription factors bind.
The DNA sequence shown above is that of the glucocorticoid response element where n represents any nucleotide. Transcription factors are often dimers of identical proteins homodimers so it is not surprising that each member of the pair needs to "see" the same DNA sequence in the same orientation.
- The DNA of many transposons is flanked by inverted repeats such as this one:
-
5' GGCCAGTCACAATGG..~400 nt..CCATTGTGACTGGCC 3'
3' CCGGTCAGTGTTACC..~400 nt..GGTAACACTGACCGG 5' - Inverted repeats at either end of retroviral gene sequences aid in inserting the DNA copy into the DNA of the host.
- Duplicated Genes.
The human Y chromosome contains 7 sets of genes — each set containing from 2 to 6 nearly-identical genes — oriented back-to-back or head-to-head; that is, they are inverted repeats like the portion shown here. (The dashes represent the thousands of base pairs that separate adjacent palindromes.)
CTCCCACAACCCATGGGATTTGTG... 3'
GAGGGTGTTGGGTACCCTAAACAC... 5'
This orientation and redundancy may help ensure that a deleterious mutation in one copy of the set can be repaired using the information in another copy of that set. All that is needed is to form a loop so that the two sequences line up side-by-side. Repairs can then be made (probably by the mechanism of homologous recombination). Here, for example, the single difference in the sequences can be eliminated (red for blue or vice versa).


