5.1: Transformation in Bacteria
- Page ID
- 4739
Bacteria have no sexual reproduction in the sense that eukaryotes do. The have no alternation of diploid and haploid generations, no gametes, and no meiosis. However, the essence of sex is genetic recombination, and bacteria do have three mechanisms to accomplish that: transformation, conjugation and transduction.
Transformation
Many bacteria can acquire new genes by taking up DNA molecules (e.g., a plasmid) from their surroundings. The ability to deliberately transform the bacterium E. coli has made possible the cloning of many genes, including human genes, and the development of the biotechnology industry. The first demonstration of bacterial transformation was done with Streptococcus pneumoniae and led to the discovery that DNA is the substance of the genes. The path leading to this epoch-making discovery began in 1928 with the work of an English bacteriologist, Fred Griffith.


The cells of S. pneumoniae (also known as the pneumococcus) are usually surrounded by a gummy capsule made of a polysaccharide. When grown on the surface of a solid culture medium, the capsule causes the colonies to have a glistening, smooth appearance. These cells are called "S" cells. However, after prolonged cultivation on artificial medium, some cells lose the ability to form the capsule, and the surface of their colonies is wrinkled and rough ("R"). With the loss of their capsule, the bacteria also lose their virulence. Injection of a single S pneumococcus into a mouse will kill the mouse in 24 hours or so. But an injection of over 100 million (100 x 106) R cells is entirely harmless.
The reason? The capsule prevents the pneumococci from being engulfed and destroyed by scavenging cells, neutrophils and macrophages, in the body. The R forms are completely at the mercy of phagocytes. Pneumococci also occur in over 90 different types: I, II, III and so on. The types differ in the chemistry of their polysaccharide capsule. Unlike the occasional shift of S -> R, the type of the organism is constant. Mice injected with a few S cells of, say, Type II pneumococci, will soon have their bodies teeming with descendant cells of the same type.
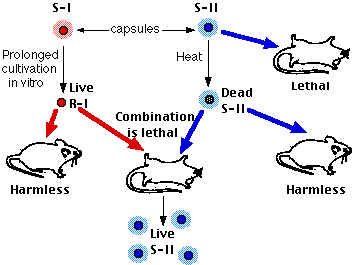
However, Griffith found that when living R cells (which should have been harmless) and dead S cells (which also should have been harmless) were injected together, the mouse became ill and living S cells could be recovered from its body. Furthermore, the type of the cells recovered from the mouse's body was determined by the type of the dead S cells. In the experiment shown above, injection of living R-I cells and dead S-II cells produced a dying mouse with its body filled with living S-II pneumococci. The S-II cells remained true to their new type. Something in the dead S-II cells had made a permanent change in the phenotype of the R-I cells. The process was named transformation.
Oswald Avery and his colleagues at The Rockefeller Institute in New York City eventually showed that the "something" was DNA. In pursuing Griffith's discovery, they found that they could bring about the same kind of transformation in vitro using an extract of the bacterial cells. They treated this extract with:
- an enzyme to destroy the polysaccharide of the capsule (S-III in their experiments)
- solvents to remove all lipids
- trypsin and chymotrypsin to destroy any residual proteins
- RNase to destroy RNA
They discovered that it did not destroy the ability of their extracts to transform the bacteria. However, treating the extracts with DNase to destroy the DNA in them did abolish their transforming activity. So DNA was the only material in the dead cells capable of transforming cells from one type to another. DNA was the substance of genes.
Although the chemical composition of the capsule is determined by genes, the relationship is indirect. DNA is transcribed into RNA and RNA is translated into proteins. The phenotype of the pneumococci — the chemical composition of the polysaccharide capsule — is determined by the particular enzymes (proteins) used in polysaccharide synthesis. Avery and his colleagues Colin MacLeod and Maclyn McCarty published their epoch-making findings on February 1, 1944. Unfortunately, the importance of their discovery was not sufficiently appreciated by scientists in general and the Nobel Committee in particular, and Avery died before their work could be honored with a Nobel Prize. (Nobel prizes are never given posthumously.)
Conjugation
Some bacteria, E. coli is an example, can transfer a portion of their chromosome to a recipient with which they are in direct contact. As the donor replicates its chromosome, the copy is injected into the recipient. At any time that the donor and recipient become separated, the transfer of genes stops. Those genes that successfully made the trip replace their equivalents in the recipient's chromosome.
Conjugation can only occur between cells of opposite mating types: the donor (or "male") carries a fertility factor (F+) and the recipient ("female") does not (F−). The fertility factor is a set of genes originally acquired from a plasmid and now integrated into the bacterial chromosome. It establishes the origin of replication for the chromosome. A portion of F is the "locomotive" that pulls the chromosome into the recipient cell and the rest of it is the "caboose".
In E. coli, about one gene gets across each second that the cells remain together. (So, it takes about 100 min for the entire genome (4377 genes) to make it. However, the process is easily interrupted so it is more likely that host genes close behind the leading F genes ("locomotive") will make it than those farther back. The "caboose" seldom makes it so failing to receive a complete F factor, the recipient cell continues to be "female".
The DNA that makes it across finds the homologous region on the female chromosome and replaces it (by a double crossover). By deliberately separating the cells (in a kitchen blender) at different times, the order and relative spacing of the genes can be determined. In this way, a genetic map — equivalent to the genetic maps of eukaryotes — can be made. However, here the map intervals are seconds, not centimorgans (cM).
Figure 5.1.4 shows the mechanism of conjugation in E. coli cells where the "male" lacks functional genes needed to synthesize the vitamin biotin and the amino acid methionine (Bio−, Met−) so these must be added to its culture medium. The "female" has those genes (Bio+, Met+) but has nonfunctional (mutant) genes that prevent it from being able to synthesize the amino acids threonine and leucine (Thr−, Leu−), so these must be added to its culture medium.
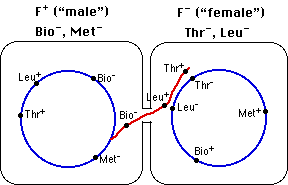
When cultured together, some female cells receive the functional Thr and Leu genes from the male donor. A double crossover enables them to replace the nonfunctional alleles. Now the cells now can grow on a "minimal" medium containing only glucose and salts.
Transduction
Bacteriophages are viruses that infect bacteria. In the process of assembling new virus particles, some host DNA may be incorporated in them. The virion head can hold only so much DNA so these viruses while still able to infect new host cells and may be unable to lyze them. Instead the hitchhiker bacterial gene (or genes) may be inserted into the DNA of the new host, replacing those already there and giving the host an altered phenotype. This phenomenon is called transduction.
The understanding of complex systems almost always has to await unraveling the details of some simpler system. You may feel that trying to find out how one type of pneumococcus could be converted into another was an exceedingly specialized and esoteric pursuit. But Avery and his coworkers realized the broader significance of what they were observing and, in due course, the rest of the scientific world did as well. By electing to work with a well-defined system: the conversion of R forms of one type into S forms of a different type, these researchers made a discovery that has revolutionized biology and medicine. Attempting to understand the workings of complex systems by first understanding the workings of their parts is called reductionism. Some scientists (and many nonscientists) question the value of reductionism. They favor a holistic approach emphasizing the workings of the complete system.
However, the record speaks for itself. From skyscrapers to moon walks, to computer chips to the advances of modern medicine, progress comes from first understanding the properties of the parts that make up the whole. The late George Wald, who won the 1967 Nobel Prize in Physiology for his discoveries of the molecular basis of detecting light, once worried that his work was overly specialized — studying not vision, not the eye, not the whole retina, not even their rods and cones, but just the chemical reactions of their rhodopsins. But he came to realize "it is as though this were a very narrow window through which at a distance one can see only a crack of light. As one comes closer, the view grows wider and wider, until finally through this same window one is looking at the universe. I think this is the way it always goes in science, because science is all one. It hardly matters where one enters, provided one can come closer....".
Significance of Genetic Recombination in Bacteria
Transformation, conjugation, and transduction were discovered in the laboratory. How important are these mechanisms of genetic recombination in nature? The completion of the sequence of the entire genome of a variety of different bacteria (and archaea) suggest that genes have in the past moved from one species to another. This phenomenon is called lateral gene transfer (LGT). The remarkable spread of resistance to multiple antibiotics may have been aided by the transfer of resistance genes within populations and even between species. Many bacteria have enzymes that enable them to destroy foreign DNA that gets into their cells. It seem unlikely that these would be needed if that did not occur in nature. In any case, these restriction enzymes have provided the tools upon which the advances of molecular biology and the biotechnology industry depend.


