1.4: Noncovalent Bonding
- Page ID
- 3744
Noncovalent Bonding
Noncovalent bonding does not involve sharing of electrons. Instead it:
- holds the two strands of the DNA double helix together (hydrogen bonds)
- folds polypeptides into such secondary structures as the alpha helix and the beta conformation
- enables enzymes to bind to their substrate
- enables antibodies to bind to their antigen
- enables transcription factors to bind to each other
- enables transcription factors to bind to DNA
- enables proteins (e.g. some hormones) to bind to their receptor
- permits the assembly of such macromolecular machinery as
- ribosomes
- actin filaments
- microtubules
- and many more
There are three principle kinds of noncovalent forces:
- ionic interactions
- hydrophobic interactions
- hydrogen bonds
Ionic Interactions
At any given pH, proteins have charged groups that may participate in binding them to each other or to other types of molecules. For example, as the figure shows, negatively-charged carboxyl groups on aspartic acid (Asp) and glutamic acid (Glu) residues may be attracted by the positively-charged free amino groups on lysine (Lys) and arginine (Arg) residues.
Ionic interactions are highly sensitive to
- changes in pH.
As the pH drops,
- H+ bind to the carboxyl groups (COO-) of aspartic acid (Asp) and glutamic acid (Glu), neutralizing their negative charge, and
- H+ bind to the unoccupied pair of electrons on the N atom of the amino (NH2) groups of lysine (Lys) and arginine (Arg) giving them a positive charge
The result: Not only does the net charge on the molecule change (it becomes more positive) but many of the opportunities that its R groups have for ionic (electrostatic) interactions with other molecules and ions are altered.
As the pH rises,
- H+ are removed from the COOH groups of Asp and Glu, giving them a negative charge (COO−), and
- H+ are removed from the NH3+ groups of Lys and Arg removing their positive charge
The result: Again the net charge on the molecule changes (it becomes more negative) and, again, many of the opportunities its R groups have for electrostatic interactions with other molecules or ions are altered.
- salt concentration
Increasing salt concentration reduces the strength of ionic binding by providing competing ions for the charged residues.
Hydrophobic Interactions
 The side chains (R groups) of such amino acids as phenylalanine and leucine are nonpolar and hence interact poorly with polar molecules like water. For this reason, most of the nonpolar residues in globular proteins are directed toward the interior of the molecule whereas such polar groups as aspartic acid and lysine are on the surface exposed to the solvent. When nonpolar residues are exposed at the surface of two different molecules, it is energetically more favorable for their two "oily" nonpolar surfaces to approach each other closely displacing the polar water molecules from between them.
The side chains (R groups) of such amino acids as phenylalanine and leucine are nonpolar and hence interact poorly with polar molecules like water. For this reason, most of the nonpolar residues in globular proteins are directed toward the interior of the molecule whereas such polar groups as aspartic acid and lysine are on the surface exposed to the solvent. When nonpolar residues are exposed at the surface of two different molecules, it is energetically more favorable for their two "oily" nonpolar surfaces to approach each other closely displacing the polar water molecules from between them.
The strength of hydrophobic interactions is not appreciably affected by changes in pH or in salt concentration.
Hydrogen Bonds
Hydrogen bonds can form whenever
- a strongly electronegative atom (e.g., oxygen, nitrogen) approaches
- a hydrogen atom which is covalently attached to a second strongly-electronegative atom
Some common examples:
- between the −C=O group and the H-N− group of nearby peptide bonds in proteins giving rise to the alpha helix and beta configuration


- Between −C=O groups and hydroxyl (H-O−) groups in
- serine and threonine residues of proteins and
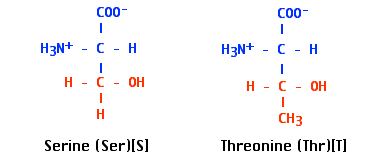
- sugars
- serine and threonine residues of proteins and
Noncovalent interactions are individually weak but collectively strong.
All three forms of noncovalent interactions are individually weak (on the order of 5 kcal/mole) as compared with a covalent bond (with its 90–100 kcal/mole of bond energy). And what strength these interactions do have requires that the interacting groups can approach each other closely (an angstrom or less). So we can conclude that all the examples given at the top of the page require:
- a substantial number of noncovalent interactions working together to hold the structures together
- a surface topography that enables substantial areas of two interacting surfaces to approach each other closely; that is, they must fit each other


