1.8: Gene Expression- Applied Example (Part 2)
- Page ID
- 73673
At the completion of this lesson you should be able to…
- Explain how a mutation in a single gene can control resistance to herbicides such as ALS inhibitors.
- Predict the impact of a specific mutation on gene expression and the eventual trait observed in a plant.
- Distinguish between the molecular and classical definitions of an allele.
- Describe the molecular basis of dominance or a lack of dominance between alleles.
This lesson describes how changes in the DNA sequence of a gene can alter the synthesis of a protein and thus influence traits such as herbicide resistance. In this lesson we will describe how changes in the gene can alter the gene expression process and influence traits in an organism. The specific example of ALS-inhibitor herbicide resistance is used to demonstrate the impact of genetic change on trait expression in a plant.
Sequence Determines Structure, Structure Determines Function
Transcription and translation are fundamental to the production of all proteins found in all living things. That is why discovering the sequence of a gene can allow geneticists to predict the amino acid sequence of the protein that gene encodes. Remember, a completely synthesized protein will be hundreds of amino acids long and thus an average gene will have two or three thousand nucleotides in its sequence. As more is learned about how amino acid sequence dictates protein function, the value of gene sequence information to biologists increases. In this lesson, we will apply our understanding of the gene expression process to understanding the genetic basis of resistance to ALS herbicides in plants.
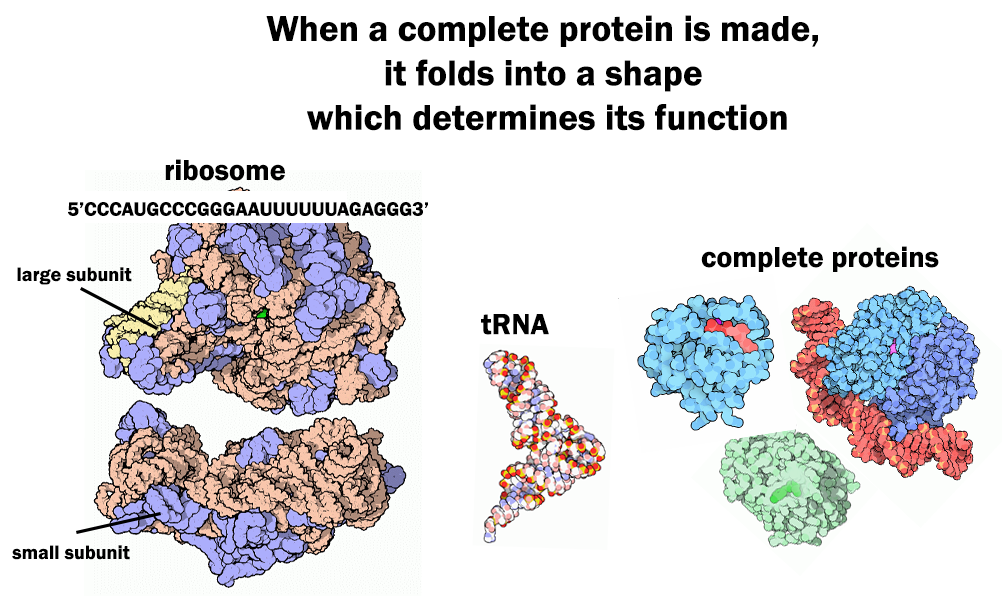
The ALS enzyme is about 800 amino acids. The specific order of amino acids in the protein chain dictates how the molecule can be folded up into a secondary structure (Fig. 1).[1] This is because each amino acid has a different chemistry that influences its interaction with other amino acids within the protein chain. The protein’s secondary structure determines the function of the protein. We can relate this principle to functional objects that are more familiar to us. Why would someone need a set of wrenches? Every tool in the set is made of the same materials. However, the specific shape and dimensions of each tool allow it to perform a specific function. The ALS enzyme functions to catalyze reactions for amino acid synthesis in the chloroplast in a plant cell. Therefore, the plant ALS enzyme structure signals the cell to transport the enzyme to the chloroplast and then allows the enzyme to catalyze the formation of a bond between amino acid precursor molecules. Clearly it was important to assemble the amino acids properly when building the ALS enzyme to allow it to function properly.
ALS and Weed Control
The function of the ALS enzyme in plants is also an interesting issue in the area of weed control. Herbicides have been discovered with sulfonyl urea or imidazolinone chemistries that will bind to ALS enzymes in plants and disrupt their catalytic functions (Figure 2). Because the herbicides bind tightly to ALS and ALS is not found in high copies in the cell, the herbicides will effectively halt the synthesis of the Leucine, Isoleucine, and Valine amino acids (Leu, Ile, Val). How will plants respond to the shortage of these three amino acids? They quit growing because they cannot make three of the twenty amino acids.
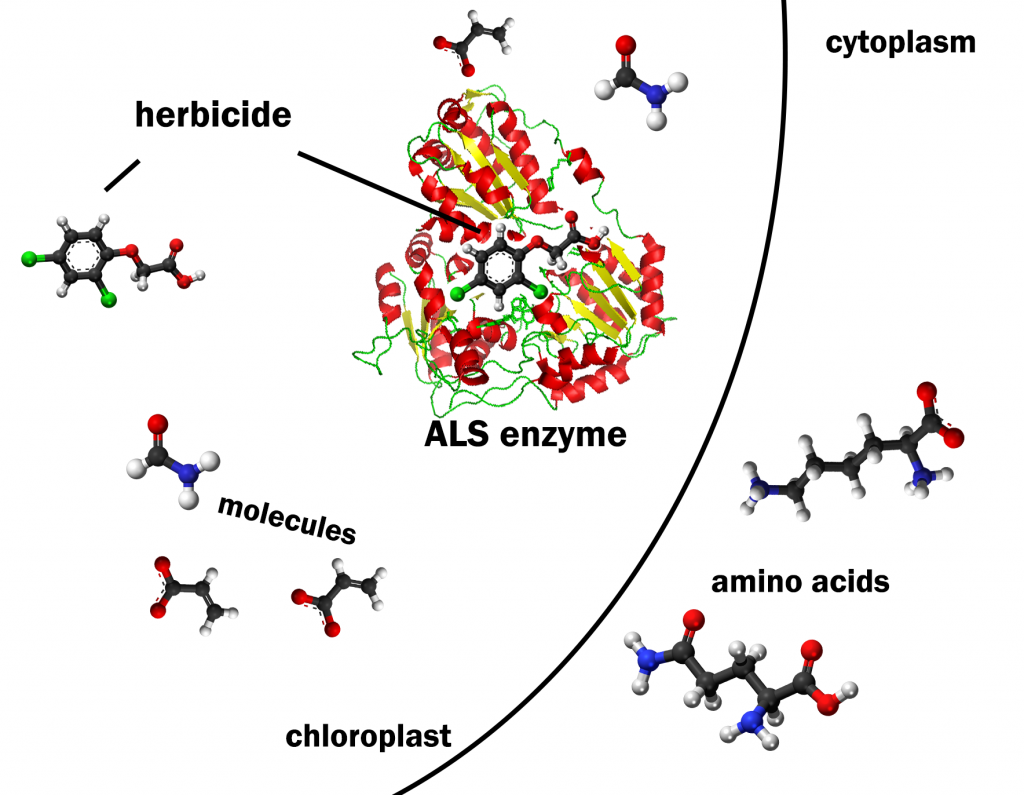
The proteins that need these amino acids will be only partially synthesized. The partial proteins will not work because they lack the structure needed for their function. The plant cells cannot grow and divide and the plant’s growth and development are halted. They will not necessarily die but they will no longer be able to compete for light and other resources if nearby plants are not affected by the ALS inhibitors. If the nearby plant that is not affected by the ALS inhibitors is a crop species such as corn or soybean, a valuable post emergence weed control strategy can be implemented.
ALS Alleles
Post emergence weed control by ALS inhibitors works in corn and soybean genotypes with altered ALS genes. The altered genes code for a slightly modified ALS enzyme which is resistant to ALS inhibitors. The molecular basis of this alteration can be determined and help us understand what alleles of a gene are. Even without molecular analysis, geneticists discovered alleles of the ALS gene that would make plants resistant to the ALS herbicides. How were these alleles discovered?
Corn breeders knew that ALS resistance did not naturally occur in their breeding lines. This discovery was easy to make. They planted thousands of different lines in short rows in the field, sprayed the field with the herbicide and observed the reaction of the lines. All of the lines tested had significant stunting. Variation did not exist in their lines so they decided to try to induce variation by mutagenesis. Pollen was collected from plants that had been exposed to a chemical mutagen. The chemical induced mistakes in DNA replication during pollen formation. The pollen was viable but carried mutant alleles. This pollen was applied to the silks of other corn plants to produce thousands of seeds. This seed was planted and the field test for ALS resistance was repeated. This time some plants resistant to stunting from ALS were observed, tagged and self-pollinated. From this experiment, the IT (immi tolerant) allele was discovered. The IT allele appeared to be dominant over the normal ALS allele because plants with one copy of the IT allele per cell (heterozygotes) had the same level of resistance as plants that had two copies of the IT allele (homozygotes). Molecular analysis of these plants was done to reveal how the IT allele was different for the original version.
The IT Allele
The IT allele of the ALS gene has a change in the coding region sequence. This was discovered by making a gene library from the DNA of homozygous IT plants and selecting a clone of the IT gene. The gene clone was sequenced and compared to the normal susceptible ALS gene from corn. A single nucleotide substitution in the IT allele was responsible for endowing a corn plant with resistance to ALS herbicides. The nucleotide substitution was in the gene coding region so it changed a codon sequence in the mRNA, coding for a different amino acid in the protein. The amino acid substitution had a minor or negligible effect on the enzyme’s ability to perform its normal functions. However, the amino acid substitution changed the site at which the ALS herbicide binds to the enzyme. The ALS herbicide will not interact with the ALS enzyme encoded by the IT allele, giving the plant resistance to the herbicide. This is an example of how a small change in a gene can have a big impact on a plant’s phenotype in the right environment.
The IR Allele, Tissue Culture Selection for Mutations
ALS alleles that confer resistance to immidazolinone herbicides have also been selected from mutations that have occurred in tissue culture cells. DNA replication does not occur 100 percent without errors so it is not surprising that natural mutations occur in cells. A mutation, though, that gives a single cell in a corn plant resistance to ALS herbicides will not render the whole plant resistant. Geneticists have used the power of tissue culture to select for these mutations at the single cell level. Hundreds of thousands of corn cells can be grown in tissue culture plates that contain the herbicide. Most of the cells will quit growing and dividing when they take up the herbicide but some cells continue to grow and form clumps of undifferentiated cells called callus. Plants regenerated from these callus cells will often express the herbicide resistance as well. The IR allele of the ALS gene was selected in this way.
This allele also has an altered coding region sequence that blocks the ALS herbicides from binding. The ALS enzyme encoded by the IR allele, however, does not provide desired levels of herbicide resistance in a heterozygous plant. The molecular mechanisms causing this difference in the IR compared to IT allele are not clear. The implication of this difference in plant breeding are significant. The IT allele can be inherited from just one parent to give the ‘Clearfield’ or “Immi” hybrid herbicide resistance. Hybrids must be homozygous for the IR allele requiring both parents be homozygous IR inbreds (Figure 3).
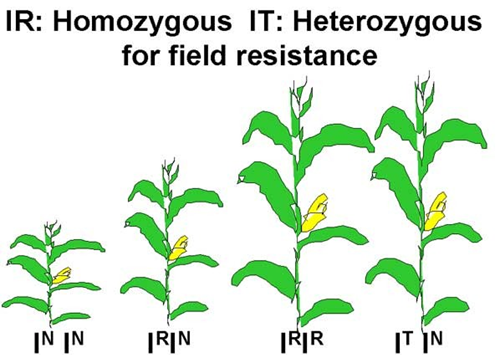
Molecular Basis of Dominance
The occurrence of the IT, IR and normal alleles of the ALS enzyme and their impact on plant phenotype provides us with an opportunity to think about dominance and lack of dominance at the molecular level. The IT allele has complete dominance to the normal allele because there is apparently no phenotype difference between homozygous dominant and heterozygous IT plants when they are sprayed with an ALS inhibitor. This suggests that in the heterozygous plants, one copy of the IT allele per cell encodes enough resistant ALS protein to drive the synthesis of sufficient quantities of Val, Leu and Ile for normal growth in the corn plant. However, the ALS enzyme encoded by the normal allele in these heterozygotes can be inhibited by the herbicide and will not contribute to amino acid synthesis.
In contrast, the IR allele has a lack of dominance to the normal allele (IN). Heterozygous plants that have one copy of the IR and one copy of the normal allele per cell do not maintain normal growth after ALS inhibitor treatment. One copy of the IR allele per cell does not encode sufficient copies of a resistant ALS enzyme to support normal growth. Instead, two copies of the IR allele per cell are needed so that all of the ALS enzyme made will not be bound by the herbicide.
The classification of the plant phenotype for characterizing possible genotypes at the ALS locus may depend on environment. In some growing environments it has been noticed that heterozygous IT hybrids will show a slight growth reduction in the sprayed plants compared to unsprayed. The difference is minor because the plants usually grow out of this difference later on and yield reduction is not a factor. This observation does emphasize that phenotype is a result of genotype and environment. Therefore, scoring traits in the proper environment will be critical to discovering the nature of gene action.
How Universal is the Genetic Code?
The table of codons used by organisms to translate mRNA into proteins is shown on the bottom of the page (Table 1). As was mentioned earlier in this lesson, the genetic code needed to be cracked one time because all organisms used the same codons to encode amino acids. As scientists began to sequence the coding regions of genes from different organisms, they discovered something called a codon preference. When you look at the codon table, you can see that the genetic code is redundant. This means that more than one codon can encode the same amino acid. This is because there are 61 codons that code for the placement of 20 different amino acids. A codon will only work in coding if a tRNA with a complementary anticodon is also found in the same cell and has the appropriate amino acid to deliver. Therefore, there could be 61 different tRNAs, one to complement each codon. Each different tRNA needs to be encoded by a different gene. If that gene is not expressed in the cell, the tRNA will not be found and a codon that needs to be complemented by that tRNA will not be complemented. In this case, the codon will act like a stop codon. The ribosome will halt translation and the protein made will be a shorter version of the intended protein. Organisms would not benefit from this situation so there is a tight complementation between what tRNAs genes are present and expressed in an organism’s cells and what codons are used to encode a specific mRNA. In this way the genetic code will have a dialect. The language is universal but certain words are used preferentially.
|
First Position |
Second Position |
Third Position |
|||
|
U |
C |
A |
G |
||
|
U |
Phe (F) |
Ser (S) |
Tyr (Y) |
Cys (C) |
U |
|
C |
|||||
|
Leu (L) |
Stop |
Stop |
A |
||
|
Stop |
Trp (W) |
G |
|||
|
C |
Leu (L) |
Pro (P) |
His (H) |
Arg (R) |
U |
|
C |
|||||
|
Gln (Q) |
A |
||||
|
G |
|||||
|
A |
Ile (I) |
Thr (T) |
Asn (N) |
Ser (S) |
U |
|
C |
|||||
|
Lys (K) |
Arg (R) |
A |
|||
|
Met (M) |
G |
||||
|
G |
Val (V) |
Ala (A) |
Asp (D) |
Gly (G) |
U |
|
C |
|||||
|
Glu (E) |
A |
||||
|
G |
|||||
Scientists are not sure why codon preferences are a part of the gene expression process in organisms. It may provide another level for the organism to control the amounts and kinds of proteins made in its cells. Recent experiences in genetic engineering of plants and animals, however, has made codon preference an important consideration. For example, scientists have put genes from a soil bacterium called Bacillus thuringiensis (Bt) into corn plant cells in order to give the corn plant the ability to make a protein that is toxic to European corn borer, a common pest to corn producers. They found that the gene would be transcribed but the mRNA would not be translated to make the desired protein. One reason was codon usage. Some of the codons the bacteria use to encode amino acids are rarely used by corn. The corn plant either lacked the tRNA to complement the codon or make the tRNA at such low levels that there were not enough copies in the cell to accommodate translation of the Bt mRNA. Therefore, the genetic engineers needed to make synthetic coding regions that substituted codons preferred by corn for those preferred by bacteria. The end result was they were able to get higher levels of the Bt protein made once these changes were made in the gene. Codon preference thus makes the genetic engineering process more challenging.
Watch this animation of the transcription process.
Watch this animation of the translation process.
Watch this video overview of Transcription and Translation.
- Graphics used in this figure were taken from The RCSB PDB "Molecule of the Month": Inspiring a Molecular View of Biology D.S. Goodsell, S. Dutta, C. Zardecki, M. Voigt, H.M. Berman, S.K. Burley (2015) PLoS Biol 13(5): e1002140. doi: 10.1371/journal.pbio.1002140 ↵


