Basic Mechanisms of Replication
- Page ID
- 317
DNA Replication is Semiconservative
We begin our investigation by describing the basic model for how nucleotides are joined in a specific order during DNA replication. By the early 1950’s, it was clear that DNA was a linear string of deoxyribonucleotides. At that point, one could postulate three different ways to replicate the DNA of a cell. First, a cell might have a DNA-synthesizing "machine" which could be programmed to make a particular string of nucleotides for each chromosome. A second possibility is that the process of replication could break the parental DNA into pieces and use them to seed synthesis of new DNA.
A third model could be proposed from the DNA structure deduced by Watson and Crick. When they described the double-helical structure of DNA in a one-page article in Nature in 1953, they included this brief statement of a third model:
"It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material."
A subsequent paper elaborated on this mechanism. The complementarity between base pairs (A with T and G with C) not only holds the two strands of the double helix together, but the sequence of one strand is sufficient to determine the sequence of the other. Hence a third possibility for a mechanism of DNA replication was clear - one parental strand could serve as atemplate directing synthesis of a complementary strand in the daughter DNA molecules. This 1953 paper is of course most famous for its description of the double-helical structure of DNA held together by base complementarity, but it is also important because the proposed structure suggested a testable model for how a particular process occurs, in this case replication.
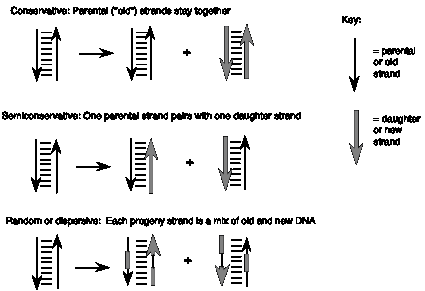
These three models make different predictions about the behavior of the two strands of the parental DNA during replication (Figure 5.1). In the first, programmed machine model, the two strands of the parental DNA can remain together, because they are not needed to determine the sequence of the daughter strands. This model of replication is called conservative: the parental DNA molecules are the same in the progeny as in the parent cell. In the second model, the each strand of the daughter DNA molecules would be a combination of old and new DNA. This type of replication is referred to as random (or dispersive). The third model, in which one strand of the parental DNA serves as a template directing the order of nucleotides on the new DNA strand, is a semiconservative mode of replication, because half of each parent duplex (i.e. one strand) remains intact in the daughter molecules.
When they were graduate students at the California Institute of Technology, Matthew Meselson and Franklin Stahl realized that they could test these three models for replication by distinguishing experimentally between old and new strands of DNA. They labeled the old or parental DNA with nucleotides composed of a heavy isotope of nitrogen (15N) by growing E. colicells for several generations in media containing [15N] NH4Cl. Ammonia is a precursor in the biosynthesis of the purine and pyrimidine bases, and hence this procedure labeled the nitrogen in the nucleotide bases in the DNA of the E. colicells with 15N. The cells were then shifted to grow in media containing the highly abundant, light isotope of nitrogen, 14N, in the NH4Cl, so that newly synthesized DNA would have a "light" density. The labeled, heavy (old) DNA could be separated from the unlabeled, light (new) DNA on a CsCl density gradient, in which the DNA bands at the position on the gradient where the concentration of CsCl has a density equal to that of the macromolecule. At progressive times after the shift to growth in [14N] NH4Cl, samples of the cells were collected, then DNA was isolated from the cells and separated on a CsCl gradient.
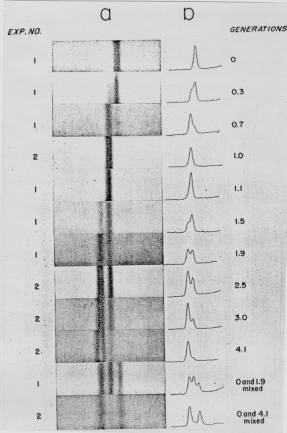
A. B.
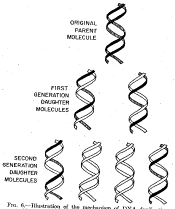
Figure 5.2. Results of the Meselson and Stahl experiment demonstrating semiconservative replication of DNA. A. The left panel (a) shows ultraviolet absorption photographs of DNA after equilibrium sedimentation in a CsCl gradient, as a function of the number of generations from the shift from media that labeled DNA with a high density (15N-labeled) to a medium in which the DNA is normal, or light density (14N-DNA). The density of the CsCl gradient increases to the right. The panel on the right (b) shows a trace of the amount of DNA along the gradient. The number of generations since the shift to the media with 14N substrates is shown at the far right. Mixing experiments at the bottom show the positions of uniformly light and heavy DNA (generations 0 and 4.1 mixed) and the mixture of those plus hybrid light and heavy DNA (generations 0 and 1.9 mixed). Parental DNA forms a band at the heavy density (15N-labeled), whereas after one generation in light (14N) media, all the DNA forms a band at a hybrid density (between heavy and light). Continued growth in light media leads to the synthesis of DNA that is only light density. B. The interpretation of the experimental results as demonstrating a semiconservative model of replication. Part A of this figure is Figure 4 and Part B is Figure 6 from M. Meselson and F. Stahl (1958) “The Replication of DNA in Escherichia coli” Proceedings of the National Academy of Sciences, USA 44:671-682.
The results fit the pattern expected for semiconservative replication (Figure 5.2). To quote from Meselson and Stahl, “until one generation time has elapsed, half-labeled molecules accumulate, while fully labeled DNA is depleted. One generation time after the addition of 14N, these half-labeled or ‘hybrid’ molecules alone are observed. Subsequently, only half-labeled DNA and completely unlabeled DNA are found. When two generation times have elapsed after the addition of 14N, half-labeled and unlabeled DNA are present in equal amounts.” A conservative mode of replication is ruled out by the observation that all the DNA formed a band at a hybrid density after one generation in the [14N] NH4Cl-containing medium. However, it is consistent with either the semiconservative or random models. As expected for semiconservative replication, half of the DNA was at a hybrid density and half was at a light density after two generations in [14N] NH4Cl-containing medium. Further growth in the 14N medium resulted in an increase in the amount of DNA in the LL band.
Exercise
Question 5.1: What data from this experiment rule out a random mode of replication?
These experiments demonstrated that each parental DNA strand is used as a template directing synthesis of a new strand during DNA replication. The synthesis of new DNA is directed by base complementarity. The enzymes that carry out replication are not programmed “machines” with an inherent specificity to synthesize a given sequence, but rather the template strand of DNA determines the order of nucleotides along the newly synthesized DNA strand (Figure 5.3).
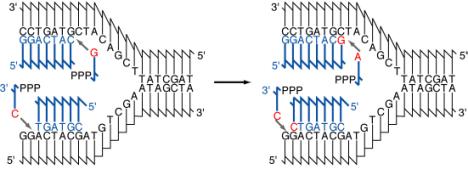
The association of a parental DNA strand with a newly synthesized DNA strand observed in this important experimental result is consistent with the use of each parental DNA strand as a template to direct the replication machinery to place nucleotides in a particular order. Watson and Crick proposed that base complementarity would guide the replication machinery to insert an A opposite a T, a T opposite an A, a G opposite a C, and C opposite a G (Figure 5.3). This was verified once the enzymes carrying out DNA synthesis were isolated, and the chemical composition of the products of replication was compared with that of the templates. These enzymes are discussed in detail later in the chapter, as will be the chemistry of the process of adding individual nucleotides to the growing DNA chain (a process called elongation). You may recall that these enzymes were also used to demonstrate the antiparallel arrangement of the DNA strands predicted by Watson and Crick (recall problem 2.5). With this understanding of how the sequence of nucleotides is specified, we can examine the types of DNA structures found during replication.


