5.4: Level 2: Contrasting spider diversity among sites to provide a basis for prioritizing conservation efforts
- Page ID
- 17375
In this part of the exercise you are provided with spider collections from 4 other forest patches. The forest patches have resulted from fragmentation of a once much larger, continuous forest. You will use the spider diversity information to prioritize efforts for the five different forest patches (including the data from the first patch which you have already classified). Here are the additional spider collections:(See Figure \(\PageIndex{1}\), Figure \(\PageIndex{2}\), Figure \(\PageIndex{3}\), and Figure \(\PageIndex{4}\))

Figure \(\PageIndex{1}\)

Figure \(\PageIndex{2}\)
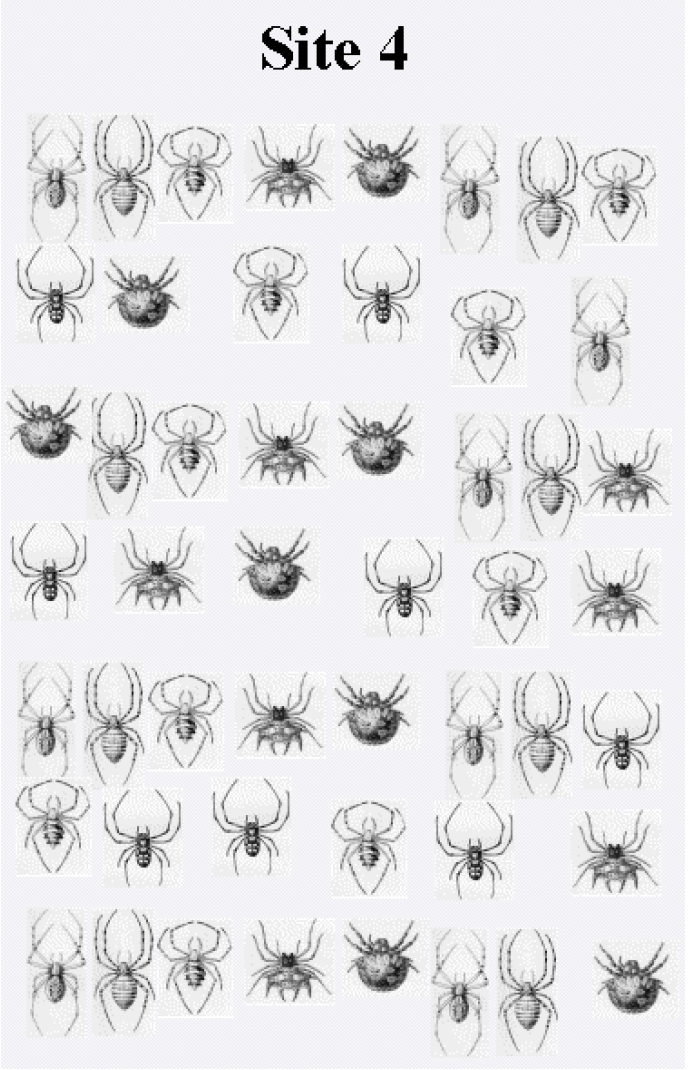
Figure \(\PageIndex{3}\)

Figure \(\PageIndex{4}\)
Again, tally how many individuals belonging to each species occur in each site's spider collection (use your classification of spiders completed for Site 1 during Level 1 of the exercise). Specifically, construct a table of species (rows) by site (columns). In the table's cells put the number of individuals of each species you found in the collection from the island. You can then analyze these data to generate different measures of community characteristics to help you to decide how to prioritize protection of the forest patches. Recall that you need to rank the patches in terms of where protection efforts should be applied, and you need to provide a rationale for your ranking.
You will find it most useful to base your decisions on three community characteristics: species richness and species diversity within each forest patch, and the similarity of spider communities between patches. Species richness is simply the tally of different spider species that were collected in a forest patch. Species diversity is a more complex concept. We will use a standard index called Simpson Reciprocal Index, \(\frac{1}{D}\) where \(D\) is calculated as follows:
\[D=∑{p_i}^2\]
where \(p_i=\) the fractional abundance of the \(i^{th}\) species on an island. For example, if you had a sample of two species with five individuals each, \(\frac{1}{0.5^2+0.5^2}=2\). The higher the value, the greater the diversity. The maximum value is the number of species in the sample, which occurs when all species contain an equal number of individuals. Because this index not only reflects the number of species present but also the relative distribution of individuals among species within a community it can reflect how balanced communities are in terms of how individuals are distributed across species. As a result, two communities may have precisely the same number of species, and hence species richness, but substantially different diversity measures if individuals in one community are skewed toward a few of the species whereas individuals are distributed more evenly in the other community.
Diversity is one thing, distinctiveness is quite another. Thus another important perspective in ranking sites is how different the communities are from one another. We will use the simplest available measure of community similarity, that is, the Jaccard coefficient of community similarity, to contrast community distinctiveness between all possible pairs of sites:
\[CC_{j}=\frac{c}{S} \]
where \(c\) is the number of species common to both communities and SS is the total number of species present in the two communities. For example, if one site contains only 2 species and the other site 2 species, one of which is held in common by both sites, the total number of species present is 3 and the number shared is 1, so 1/3=331/3=33%. This index ranges from 0 (when no species are found in common between communities) to 1 (when all species are found in both communities). Calculate this index to compare each pair of sites separately, that is, compare Site 1 with Site 2, Site 1 with Site 3, …, Site 4 with Site 5 for 10 total comparisons. You might find it useful to determine the average similarity of one community to all the others, by averaging the \(CC_j\) values across each comparison a particular site is included.
Once you have made these calculations of diversity (species richness and Simpson's Reciprocal Index) you can tackle the primary question of the exercise: How should you rank these sites for protection and why? Making an informed decision requires reconciling your analysis with concepts of biological diversity as it pertains to diversity and distinctiveness. What do you recommend?


