25.2: Current Research Directions
- Page ID
- 41086
This page is a draft and is under active development.
Encoding functionality in DNA is one way synthetic biologists program cells. As the price of sequencing and synthesis of DNA continues to decrease, coding DNA strands has become more feasible. In fact, the number of base pairs that can be synthesized per US$ has increased exponentially, akin to Moore’s Law.
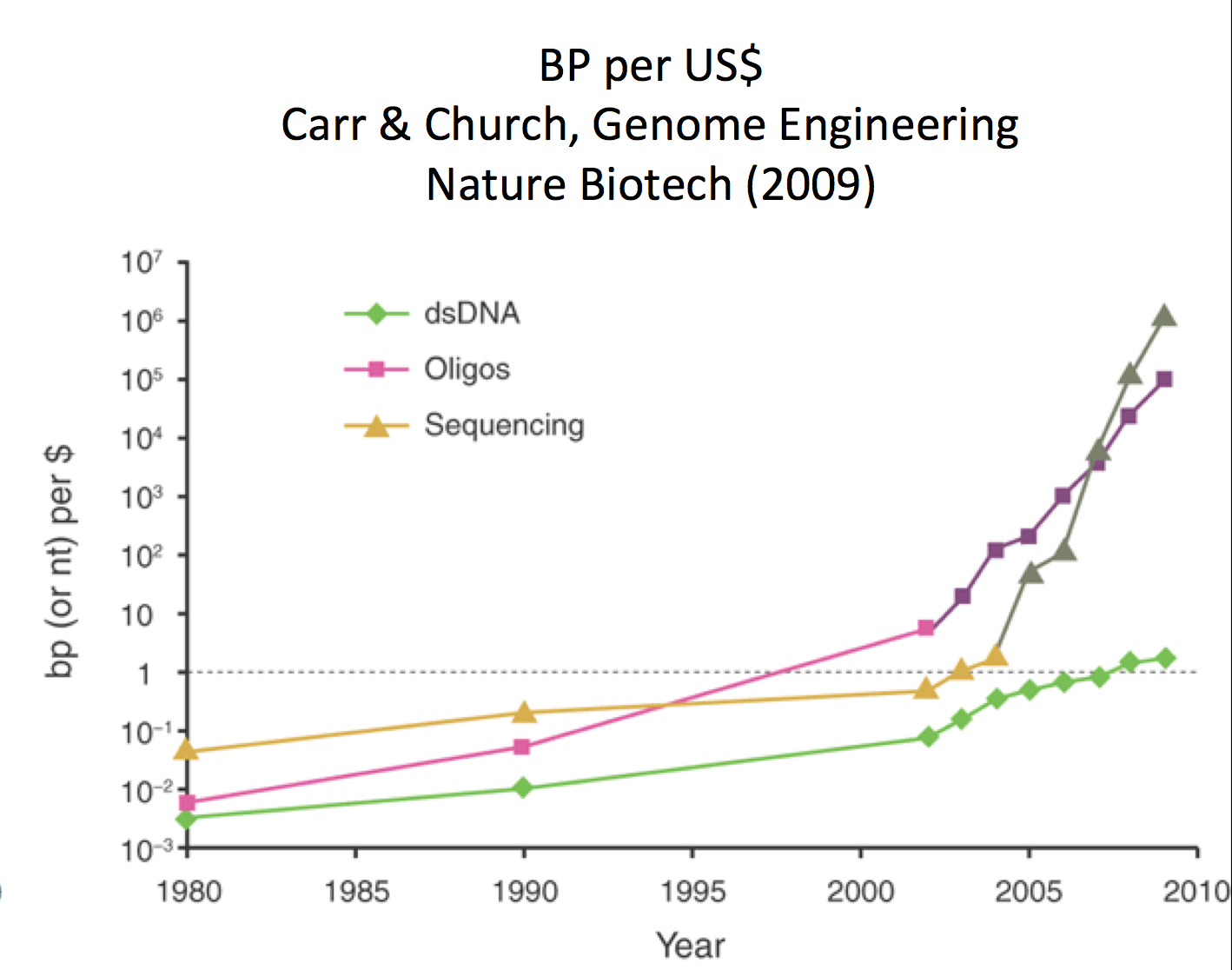
Source: Carr, Peter A. and George M. Church. "Genome Engineering."
Nature Biotechnology 27, no. 12 (2009): 1151-62
Figure 26.4: Cost of synthesizing a base pair versus US dollar
This has made the process of designing, building, and testing biological circuits much faster and cheaper. One of the major research areas in synthetic biology is the creation of fast, automated synthesis of DNA molecules and the creation of cells with the desired DNA sequence. The goal of creating a such a system is speeding up the design and debugging of making a biological system so that synthetic biological systems can be prototyped and tested in a quick, iterative process.
Synthetic biology also aims to develop abstract biological components that have standard and well-defined behavior like a part an electrical engineer might order from a catalogue. To accomplish this, the Registry of Standard Biological Parts (partsregistry.org) [4] was created in 2003 and currently contains over 7000 available parts for users. The research portion of creating such a registry includes the classification and description of biological parts. The goal is to find parts that have desirable characteristics such as:
Orthogonality Regulators should not interfere with each other. They should be independent.
Composability Regulators can be fused to give composite function.
Connectivity Regulators can be chained together to allow cascades and feedback.
Homogeneity Regulators should obey very similar physics. This allows for predictability and efficiency.
Synthetic biology is still developing, and research can still be done by people with little background in the field. The International Genetically Modified Machine (iGEM) Foundation (igem.org) [3] created the iGEM competition where undergraduate and high school students compete to design and build biological systems that operate within living cells. The student teams are given a kit of biological parts at the beginning of the summer and work at their own institutions to create biological system. Some interesting projects include:
Arsenic Biodetector The aim was to develop a bacterial biosensor that responds to a range of arsenic concentrations and produces a change in pH that can be calibrated in relation to arsenic concentration. The team’s goal was to help many under-developed countries, in particular Bangladesh, to detect arsenic contamination in water. The proposed device was intended be more economical, portable and easier to use in comparison with other detectors.
BactoBlood The UC Berkeley team worked to develop a cost-effective red blood cell substitute constructed from engineered E. coli bacteria. The system is designed to safely transport oxygen in the bloodstream without inducing sepsis, and to be stored for prolonged periods in a freeze-dried state.
E. Chromi The Cambridge team project strived to facilitate biosensor design and construction. They designed and characterised two types of parts - Sensitivity Tuners and Colour Generators – E. coli engineered to produce different pigments in response to different concentrations of an inducer. The availability of these parts revolutionized the path of future biosensor design.

