21.5: Structural Properties of Networks
- Page ID
- 41048
Much of the early work on networks was done by scientists outside of biology. Physicists looked at internet and social networks and described their properties. Biologists observed that the same properties were also present in biological networks and the field of biological networks was born. In this section we look at some of these structural properties shared by the different biological networks, as well as the networks that arise in other disciplines as well.
Degree distribution
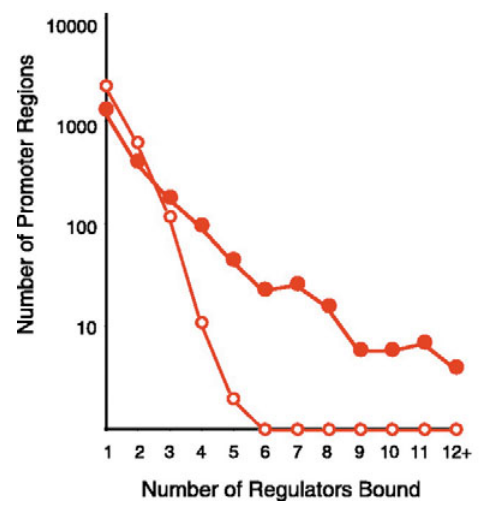
source unknown. All rights reserved. This content is excluded from our Creative Commons license. For more information, see ocw. mit.edu/help/faq-fair-use/.
In a network, the degree of a node is the number of neighbors it has, i.e., the number of nodes it is connected to by an edge. The degree distribution of the network gives the number of nodes having degree d for each possible value of d = 1, 2, 3, . . . . For example figure 21.3 gives the degree distribution of the S. cerevisiae gene regulatory network. It was observed that the degree distribution of biological networks follow a power law, i.e., the number of nodes in the network having degree d is approximately cd where c is a normalization constant and is a positive coefficient. In such networks, most nodes have a small number of connections, except for a few nodes which have very high connectivity.
This property –of power law degree distribution– was actually observed in many different networks across different disciplines (e.g., social networks, the World Wide Web, etc.) and indicates that those networks are not “random”: indeed random networks (constructed from the Erd ̋os-Renyi model) have a degree distribution that follows a Poisson distribution where almost all nodes have approximately the same degree and nodes with higher or smaller degree are very rare [6] (see figure 21.4).
Networks that follow a power law degree distribution are known as scale-free networks. The few nodes in a scale-free network that have very large degree are called hubs and have very important interpretations. For example in gene regulatory networks, hubs represent transcription factors that regulate a very large number of genes. Scale-free networks have the property of being highly resilient to failures of “random” nodes, however they are very vulnerable to coordinated failures (i.e., the network fails if one of the hub nodes fails, see [1] for more information).
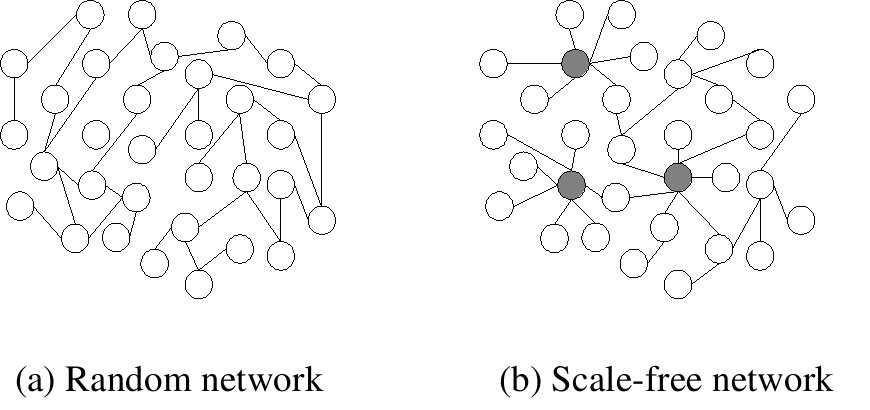_Random_network.png?revision=1)
Creative Commons license. For more information, see http://ocw.mit.edu/help/faq-fair-use/.
(a) Scale-free graph vs. a random graph (figure taken from [10]) .
(a) Scale-free graph vs. a random graph (figure taken from [10]) .
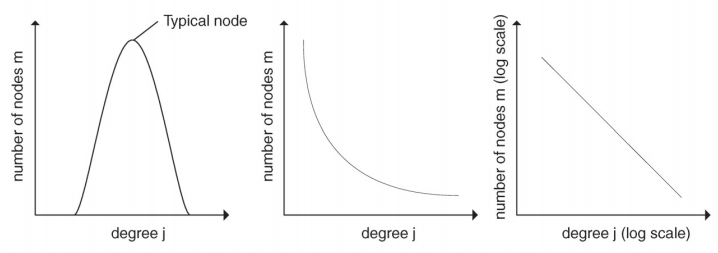
Vieweg Verlag. All rights reserved. This content is excluded from our Creative
Commons license. For more information, see http://ocw.mit.edu/help/faq-fair-use/.
Source: Hein, Oliver, et al. "Scale-free Networks." Wirtschaftsinformatik 48, no. 4
(2006): 267-75.
Figure 21.4: Scale-free vs. random Erd ̋os-Renyi networks
In a regulatory network, one can identify four levels of nodes:
- Influential, master regulating nodes on top. These are hubs that each indirectly control many targets.
- Bottleneck regulators. Nodes in the middle are important because they have a maximal number of direct targets.
- Regulators at the bottom tend to have fewer targets but nonetheless they are often biologically essential!
- Targets.
Network motifs
Network motifs are subgraphs of the network that occur significantly more than random. Some will have interesting functional properties and are presumably of biological interest.
Figure 21.5 shows regulatory motifs from the yeast regulatory network. Feedback loops allow control of regulator levels and feedforward loops allow acceleration of response times among other things.
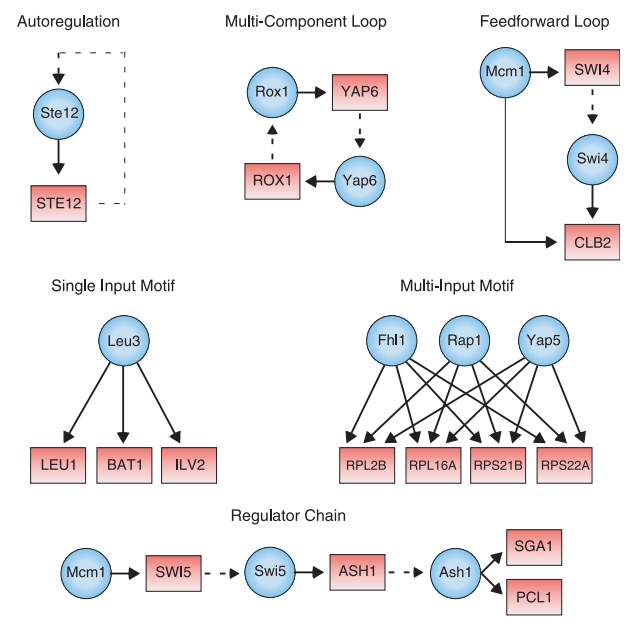 Figure 21.5: Network motifs in regulatory networks: Feed-forward loops involved in speeding-up response of target gene. Regulators are represented by blue circles and gene promoters are represented by red rectangles (figure taken from [4])
Figure 21.5: Network motifs in regulatory networks: Feed-forward loops involved in speeding-up response of target gene. Regulators are represented by blue circles and gene promoters are represented by red rectangles (figure taken from [4])

