21.4: Application of Networks
- Page ID
- 41047
Using linear regression and regression trees, we will try to predict expression from networks. Using collective classification and relaxation labeling, we will try to assign function to unknown network elements.
We would like to use networks to:
- predict the expression of genes from regulators.
In expression prediction, the goal is to parametrize a relationship giving gene expression levels from regulator expression levels. It can be solved in various manners including regression and is related to the problem of finding functional networks.
- predict functions for unknown genes.
Overview of Functional Models
One model for prediction is a conditional gaussian: a simple model trained by linear regression. A more complex prediction model is a regression tree trained by nonlinear regression.
Conditional Gaussian Models
Conditional gaussian models predict over a continuous space and are trained by a simple linear regression to maximize likelihood of data. They predict targets whose expression levels are means of gaussians over regulators.
Conditional gaussian learning takes a structured, directed net with targets and regulating transcription factors. You can estimate gaussian parameters,μ, from the the data by finding parameters maximizing likelihood - after a derivative, the ML approach reduces to solving a linear equation.
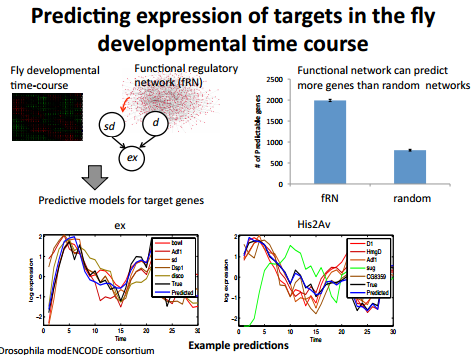
From a functional regulatory network derived from multiple data sources 6,Dr, Roy trained a gaussian model for prediction using time course expression data and tested it on a hold-out testing set. In comparisons to predictions by a modle trained from a random network, found out that the network predicted substantially better than random.
The linear model used makes a strong assumption on linearity of interaction. This is probably not a very accurate assumption to make but it appears to work to some extent with the dataset tested.
Regression Tree Models
Regression tree models allow the modeler to use a multimodal distribution incorporating nonlinear dependencies between regulator and target gene expression. The final structure of a regression tree describes expression grammar in terms of a series of choices made at regression tree nodes. Because targets can share regulatory programs, notions of recurring motifs may be incorporated. Regression trees are rich models but tricky to learn. regression trees in predicting expression
In practice, prediction works its way down a regression tree given regulator expression levels. Upon reaching the leaf nodes of the regression tree, a prediction for gene expression is made.
Functional Prediction for Unannotated Nodes
Given a network with an incomplete set of labels, the goal of function annotation is to predict labels for unknown genes. We will use methods falling under the broad category of guilt by association. If we know nothing about a node but that its neighbors are involved in a function, assign that function to the unknown node.
Association can include any notion of network relatedness discussed above such as co-expression, protein- protein interactions and co-regulation. Many methods work, two will be discussed: collective classification and relaxation classification; both of which work for regulatory networks encoded as undirected graphs.
Collective Classification
View functional prediction as a classification problem: Given a node, what is its regulatory class?.
source unknown. All rights reserved. This content is excluded from our Creative
Commons license. For more information, see http://ocw.mit.edu/help/faq-fair-use/.
In order to use the graph structure in the prediction problem, we capture properties of the neighborhood of a gene in relational attribute. Since all points are connected in a network, data points are no longer inde- pendently distributed - the prediction problem becomes substantially harder than a standard classification problem.
Iterative classification is a simple method with which to solve the classification problem. Starting with an initial guess for unlabeled genes it infers labels iteratively, allowing changed labels to influence node label predictions in a manner similar to gibbs sampling7
Relaxation labeling is another approach originally developed to trac terrorist networks. The model uses a suspicion score where nodes are labeled with a suspiciousness according to the suspiciousness of its neighbors. The method is called relaxation labeling because it gradually settles on to a solution according to a learning parameter. It is another instance of iterative learning where genes are assigned probabilities of having a given function.
Regulatory Networks for Function Prediction
For pairs of nodes, compute a regulatory similarity – the interaction quantity – equal to the size of the intersection of their regulators divided by the size of their union. Having this interaction similarity in the form of an undirected graph over network targets, can use clusters derived from a network in final functional classification.
The model is successful in predicting invaginal disk and neural system development. The blue line in Fig. 21.2a shows the score of every gene predicting its participation in neural system development.
Co-expression an co-regulation can be used side by side to augment the set of genes known to particiapte in neural system development.
6data sources included chromatin, physical binding, expression, motif
7see the previous lecture by Manolis describing motif discovery


