12.5: Long non-coding RNAs in Epigenetic Regulation
- Page ID
- 40989
Let’s examine human skin as an example of long non-coding RNAs being used in epigenetic regulation. Human skin is huge, in fact it is the largest organ by weight in the body. It is intricate, with specialized features, and it is constantly regenerating to replace old dead cells with new ones. The skin must be controlled so hair only grows on the back of your hand rather than on your palm. Moreover, these boundaries cannot change and are maintained ever since birth.
The skin in all parts of the body is composed of an epithelial layer and a layer of connective tissue made up of cells called fibroblasts. These fibroblasts secrete cytokine signals that control the outer layer, determining properties such as the presence or absence of hair. Fibroblasts all around the body are identical except for the specific epigenetic folding that dictates what type of skin will be formed in a given location. Based on whether the skin is distal or proximal, interior or exterior, posterior or anterior, a different set of epigenetic folds will determine the type of skin that forms.

Figure 12.9: Human fibroblasts specialize via epigenetic regulation to form different skin types based on their location within the body. Research has found that the type of skin in the hands shares a remarkably similar epigenetic signature to the skin in the feet, which is also distally located.
It has been found that specific HOX genes delineate these anatomical boundaries during development. Just by looking at the human HOX genetic code, one can predict where a cell will be located. Using ChIP- on-chip (chromatin immunoprecipitation microarrays) diamteric chromatin domains have been found among these HOX genes. In the figure below, we can see a clear boundary between the chromatin domains of a cell type located proximally and another located distally. Not only is this boundary precise, but it is maintained across trillions of skin cells.

Figure 12.10: Two skin cell types are analyzed for their chromatin domains. There exists a clear boundary between the lung cell type which is proximal to the body, and the foot cell type which is distal to the body.
HOTAIR or HOX transcript antisense intergenic RNA has been investigated as possible RNA regulator that keeps these boundary between the diametric domains in chromatin. When HOTAIR was knocked out in the HOXC locus, it was hypothesized that the chromatin domains might slip through into one another. While it was found that this HOTAIR did not directly affect the epigenetic boundary, researchers did find evidence of RNA based genomic cross talk. The HOTAIR gene affected a different locus called HOXD.
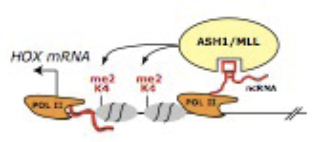
Figure 12.11: Polycomb, a protein that can remodel chromatin so that epigenetic silencing of genes can take place, may be regulated by non-coding RNA such as HOTAIR.
Through a process of ncRNA dependent Polycomb repression, the HOTAIR sequence can control epige- netic regulation. Plycomb is a portein that puts stop marks on the tails of histones so that they can cause specific folds in the genetic material. On their own histones, are undirected, so it is necessary for some mechanism to dictate how they attach to the genome. This process of discovery has led to great interest in the power of long intergenic non-coding RNAs to affect epigenetic regulation.


