9.1: Prokaryotic Transcriptional Regulation
- Page ID
- 16142
Unlike multicellular organisms, in which most cells are in a tightly regulated internal environment, most prokaryotic cells are constantly responding to changing conditions in their immediate environment, such as changes in salt concentration, temperature, acidity, or nutrient availability. Because these organisms must respond quickly, the lifetime of an RNA is kept short, on the order of several minutes - so gene products that are not useful in the new conditions do not waste resources. For the same reason, initiation of new transcription must also occur very quickly - so that gene products that are needed to stabilize the cell in the new conditions are rapidly available. A fast and efficient control system is needed, and in prokaryotes, this means that the controls on transcription are simple activators and repressors. For some genes, both may be used for regulation, while for others, only one is needed to change from a default state of expression or non-expression.
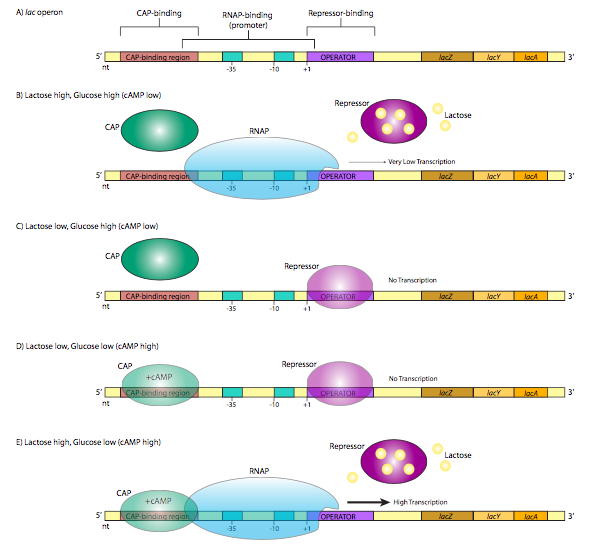
A classic example of repressor control of gene expression, the lac operon, also illustrates another method by which bacteria may control the expression of genes. An operon is a group of genes whose products participate in the same metabolic pathway, and are transcribed under the control of a single promoter. The lac operon consists of three genes (lacZ, lacY, lacA) that participate in the catabolism of the disaccharide, lactose. LacZ is β-galactosidase, an enzyme that cleaves lactose into galactose and glucose. LacY is β-galactoside permease, which transports lactose from the extracellular environment into the cell. Both are required for lactose catabolism. Oddly, lacA is not absolutely required for lactose metabolism, but its function is related to the other two: it is a β-galactoside transacetylase that transfers acetyl groups from acetyl-CoA to lactose. All three are translated (they retain their individual start and stop codons for translation, not to be confused with the start and stop of transcription) from a single transcript. Of particular interest with respect to the regulation of this transcription is the structure of the promoter region. Note that in addition to the expected σ70 promoter upstream of the start site, there is another control sequence on each side of the start site (Figure \(\PageIndex{1}\)A).
Note that the helix-turn-helix (HTH) motif, which is common in bacterial DNA-binding proteins, is not the same thing as the helix-loop-helix DNA-binding proteins that are used in many eukaryotic systems. An elaboration of the basic HTH motif, known as the winged helix motif, is also found in a variety of prokaryotic DNA- binding proteins.
The operator is a sequence of DNA that lies between the promoter and the start site. It is recognized by the lac repressor, a DNA binding protein with a helix-turn-helix motif. In the absence of lactose (Figure \(\PageIndex{1}\)C), the lac repressor has a high affinity for the operator sequence and binds tightly, obstructing the start site and forming a physical “roadblock” to transcription by preventing the RNA polymerase from moving forward from the promoter. This makes sense physiologically because the cell is more efficient metabolizing glucose, and if there is no lactose around, then it is a waste of resources to make enzymes that metabolize it. However, what if there is suddenly an abundance of lactose in the environment? As the lactose is taken into the cell, intracellular levels rise, and now enzymes are needed to utilize this new food source. The lactose actually turns on the expression of enzymes that will metabolize it! Specifically, the lactose binds to the lac repressor protein (4 lactose binding sites), which causes a conformational change that releases it from the operator sequence (Figure \(\PageIndex{1}\)B). Now an RNA polymerase that attaches at the lac operon promoter can proceed to transcribe the message unhindered, producing RNA and subsequently proteins that are used to break down the lactose. This continues as long as there is abundant lactose in the cell. As the lactose levels drop, repressor proteins are no longer bound by lactose, and can once again bind the operator and inhibit expression of the operon once again. For now, ignore the CAP protein in Figure \(\PageIndex{1}\), and parts D and E. We’ll come back to that. The lac operon is an example of an inducible operon, in which the native state is “off” and the introduction of and inducer (in this case lactose) will bind the repressor and turn the operon “on”.
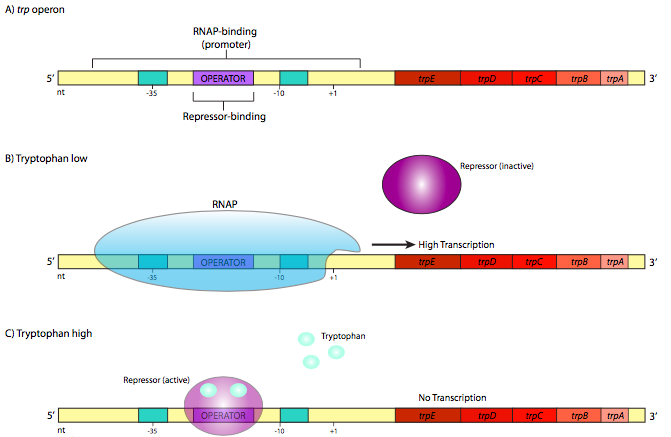
In contrast, there are also operons with the reverse mechanism. An example of one such repressible operon is the trp operon (Figure \(\PageIndex{2}\)). This operon contains ve genes that are involved in the synthesis of the amino acid tryptophan: trpE and trpD, which together encode the subunits of anthranilate synthetase, trpC, which encodes N-(5’- phosphoribosyl)-anthranilate isomerase, and trpB and trpA, which each encode sub-units of tryptophan synthetase. The trp repressor is larger and more complex than the lac repressor, but it also utilizes a helix-turn-helix DNA-binding motif.
However, it differs in a crucial aspect. In its native form, it does not bind to the opera- tor sequence. It only binds to the operator after it has first bound tryptophan (two molecules of trp bind to one repressor). This is the opposite of the lac repressor, but when considering the physiological function of these genes, this should make perfect sense. As long as there is no tryptophan, the operator is unbound, allowing the RNA polymerase to transcribe the genes needed to make tryptophan (Figure \(\PageIndex{2}\)B). When enough tryptophan has accumulated in the cell, some of the “extra” tryptophan binds to the trp repressor, which activates it and allows it to bind to the operator (Figure \(\PageIndex{2}\)C). When this happens, the RNAP cannot reach the start site, and resources are not wasted transcribing genes for enzymes that make something the cell already has a lot of.
Let us now return to the lac operon in Figure \(\PageIndex{1}\). It turns out that even when the operon is induced by the presence of lactose, the rate of transcription is low. The limitation is not from the repressor - that has been removed as described above (Figure \(\PageIndex{1}\)B). Instead, the low expression is due to a low-affinity promoter. This is true not just of the lac operon, but also other non-glucose-pathway sugar-catabolism genes. There is a simple explanation: even if there are abundant alternate sugars available (e.g. lactose), if there is glucose available, it is the cell’s most efficient and preferred pathway for energy production, and the production of enzymes for other pathways would be an inefficient use of resources. So, when and how is the lac operon really turned on?
The answer lies in a CAP, catabolite gene activator protein, also known as CRP, or cAMP receptor protein. It is a small homodimeric DNA binding protein that binds to a sequence that overlaps the 5’ side of the promoter. In the presence of cAMP, which binds to the protein, CAP has a high affinity for the DNA recognition sequence, and binds to it (Figure \(\PageIndex{1}\)E). The protein then helps to recruit the RNAP to the promoter site, binding directly to the C-terminal domain of the RNAP a subunit to increase the affinity of the polymerase for the promoter sequence to overcome a weak promoter.
What does cAMP have to do with this? When there is abundant extracellular glucose, there is little cAMP. The enzyme that synthesizes cAMP, adenylate cyclase, is negatively regulated by glucose transport. However, when there is little environmental glucose, adenylate cyclase is more active, makes cAMP, which binds CAP, and leads to robust production of lactose catabolism enzymes. CAP is an example of an activator that can control gene expression in a positive direction.
In E. coli, cAMP levels are not directly tied to intracellular glucose levels or glucose metabolism. Rather, cAMP levels are altered by glucose transport through a phosphoenolpyruvate-dependent phosphotransferase system (PTS), part of which is de-phosphorylated (the crr gene product, also known as EIIA) when glucose is moved inward. The phosphorylated EIIA~P is an activator of adenylate cyclase. So, as glucose moves into the cell, cAMP levels drop due to inactive adenylate cyclase.
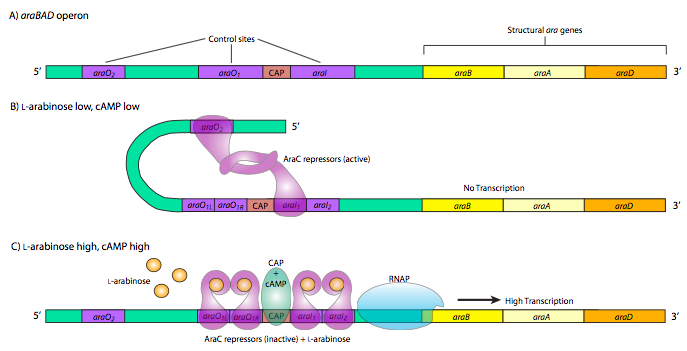
The last, and most complicated example of prokaryotic metabolic gene control is the araBAD operon. This operon produces enzymes used for the catabolism of the 5-carbon sugar, L-arabinose. The interesting thing about this operon is the presence of both positive and negative control elements that are used by the same control protein, araC. When there is little or no arabinose, the araC binds to the operator sequences araO2 and araI1. The two araC proteins then interact, which causes the DNA to loop around preventing RNAP from binding to the promoter and transcribing araBAD. Furthermore, this operon is also under the control of CAP, and the double araC loop structure also prevents CAP from binding. However, when there is plentiful arabinose, araC repres- sors bind the arabinose and then interact differently, still forming dimers, but now in a different conformation that leads to binding of araO1L and araO1R together as well as araI1 and araI2. The arabinose-bound araC at the araI sites interact with RNAP and together with CAP promote strong activation of araBAD expression.
Not all operons are concerned with coordinating metabolic activities. An important non-metabolic operon in E. coli is the LexA/ RecA SOS response operon, which contains genes that are involved in DNA repair. The SOS repair system is invoked to allow DNA replication to continue through areas of damaged DNA, but with the penalty of low fidelity. One of the gene products of this operon, RecA, is important in recognizing and repairing damage caused by UV light. It also functions as a regulator of the LexA repressor protein. LexA is actually a repressor for multiple SOS operons, binding to a common operator sequence upstream of each gene/operon. It is activated when RecA, upon detecting DNA damage, undergoes a conformational shift and activates protease activity, which then cleaves LexA, allowing transcription from the SOS genes/operons.
SOS repair is error-prone because when the replisome encounters bulky damage, it undergoes “replication fork collapse” in which the DNA polymerase III units are released. The replacement, or bypass, polymerases, Pol IV (dinB), and Pol V (umuDC), do not have 3’—5’ proofreading exonuclease activity. Misincorporation of G opposite thymine dimers occurs at about half the rate of proper A incorporation, and generally, the bypass polymerases are about 1000 times more error-prone than Pol II or Pol I.


