8.4: Post-Transcriptional Processing of RNA
- Page ID
- 16138
The first of the post-transcriptional events is 5’ end capping. Once the 5’ end of a nascent RNA extends free of the RNAP II approximately 20-30 nt, it is ready to be capped by a 7-methylguanosine structure. This 5’ “cap” serves as a recognition site for transport of the completed mRNA out of the nucleus and into the cytoplasm.
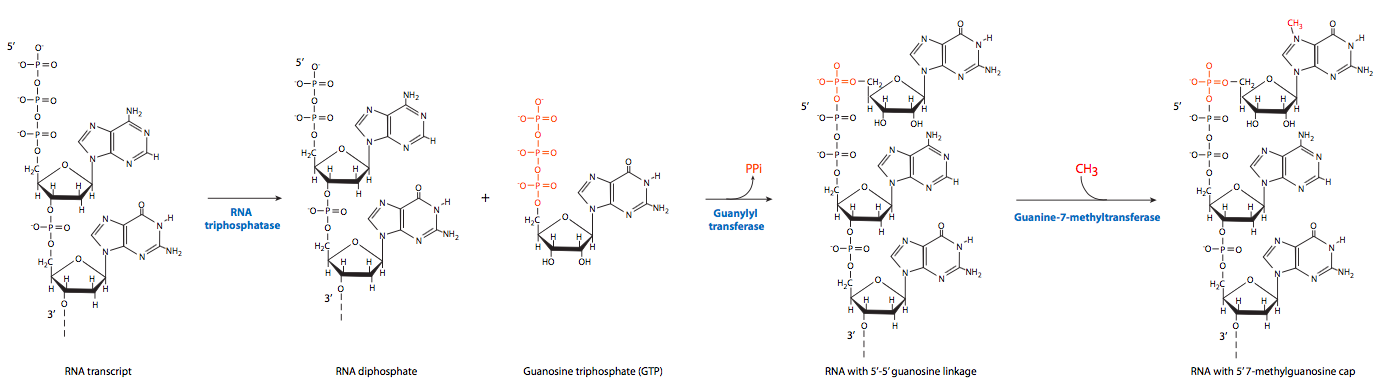
The process actually involves three steps. First, RNA triphosphatase removes the 5’-terminal triphosphate group. Guanylation by GTP is catalyzed by capping enzyme, forming an unusual 5’-5’ “backward” bond between the new guanine and the first nucleotide of the RNA transcript. Finally, guanine-7-methyltransferase methylates the newly attached guanine.
On the opposite end of the RNA, on the free 3’-OH, polyadenylation occurs. As noted previously, an enzyme complex that docks to a site on the CTD tail of RNAP II cleaves a portion of the 3’ end near an AAUAAA recognition sequence and then serially adds a large number of adenine residues. The poly(A) tail is not required for translation, but it has an effect on the stability of transcripts in the cytoplasm. As mRNA molecules stay in the cytoplasm longer, the poly(A) tail is gradually removed. Once the poly(A) tail is gone, the mRNA will soon be destroyed. mRNA molecules with longer poly(A) tails are generally longer-lived in the cytoplasm than those with shorter tails, but there is currently no evidence for a directly proportional effect.
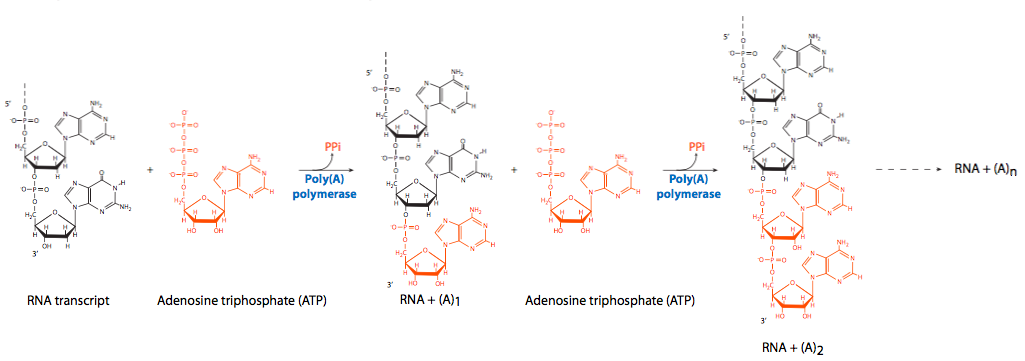
Although the enzyme that cleaves the primary transcript in preparation for polyadenylation has not been identified, two nonenzymatic factors, the excitingly-named cleavage factor I (CFI) and cleavage factor II (CFII) have been implicated. The serial adenylation comes from the activity of poly(A) polymerase (PAP) in conjunction with CPSF (cleavage and polyadenylation speci city factor), which binds to the RNA. PAP itself has relatively poor affinity for RNA. As with other nucleic acid polymerases, it adds new nucleotides onto the free 3’-OH of the pre-existing chain. To encourage processivity (continuous polymerization) poly(A) binding protein II (PABII) joins the polyadenylation complex, and is involved in controlling the final length of the poly(A) tail. It should be noted that PABII is a nuclear protein and should not be confused with PABP (poly(A) binding protein) which binds to mRNA molecules in the cytoplasm and plays a role in protecting them from nuclease attack.
The third and most complicated modification to newly-transcribed eukaryotic RNA is splicing. Unlike prokaryotic RNA, which is a continuously translatable coding region immediately as it comes out of the RNA polymerase, most eukaryotic RNAs have interrupted coding regions. Splicing is the process by which the non-coding regions, known as introns, are removed, and the coding regions, known as exons, are connected together. In some RNAs, this can happen autonomously, with part of the RNA acting as an enzymatic catalyst for the process. This requires that the RNA have a specific secondary and tertiary structure, bringing the two exons close together while looping out the intron. It was the study of this phenomenon that led to the discovery of ribozymes, which are enzymes made of RNA.
Until the discovery of ribozymes, it had been assumed that only the enzymes could only be generated with the diversity of structures possible with the amino acids in proteins.
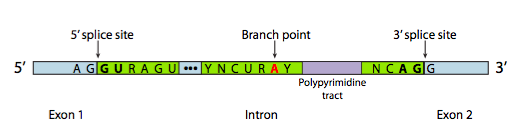
In most cases, however, splicing is carried out by a multi-subunit protein complex known as the spliceosome. Whether it is self-spliced or by spliceosome, there are three main sequence components needed to define an intron that is going to be spliced out (Figure \(\PageIndex{8}\)). There is a 5’ splice site with the consensus sequence AG|GUAAGU. There is a 3’ splice site that starts with an 11-nucleotide polypyrimidine tract followed by NCAG|G. And somewhere in between the two, there is a branchpoint adenine, typically within a YNCURAY sequence (Y is a pyrimidine, N is any nucleotide, R is a purine). Splicing is actually a set of two sequential transesterification reactions, and requires physical proximity of the reactive sites by bending and looping of the RNA, either autonomously or around protein factors know as snRNPs (pronounced “snurps”). SnRNPs is an acronym for small nuclear ribonucleoproteins. They contain both a protein and a small nuclear RNA (snRNA) component; the latter helps with sequence recognition. Examination of the structure of the snRNA part of these spliceosome snRNPs shows that they are very similar to the shapes taken by the RNA transcript itself in cases of self-splicing. Keeping that in mind, much of the following description of spliceosome-mediated splicing happens in self-splicing as well.
Although the snRNPs are the primary components of the spliceosome, a variety of other splicing factors also play a role. The most prominent are U2AF (U2-associated factor, which binds to the polypyrimidine tract, and SF1 (splicing factor 1, aka branch- point protein BPP) which binds to consensus sequence near the branchpoint. Together they help to properly position the U2 snRNP. There are also a variety of other less-studied splicing factors from the SR protein family (C-terminal Serine-Arginine binding motif) and the hnRNP (heterogenous nuclear ribonucleo- protein) families that act to recruit the primary members of the spliceosome to their proper locations.
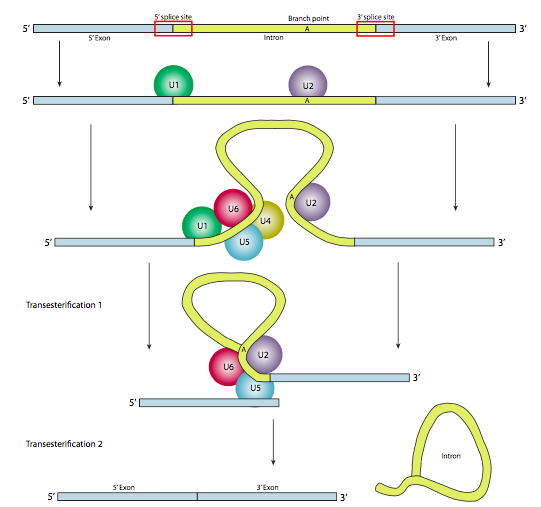
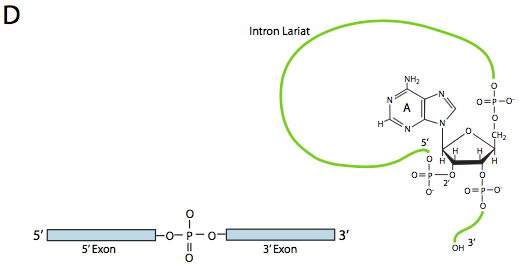
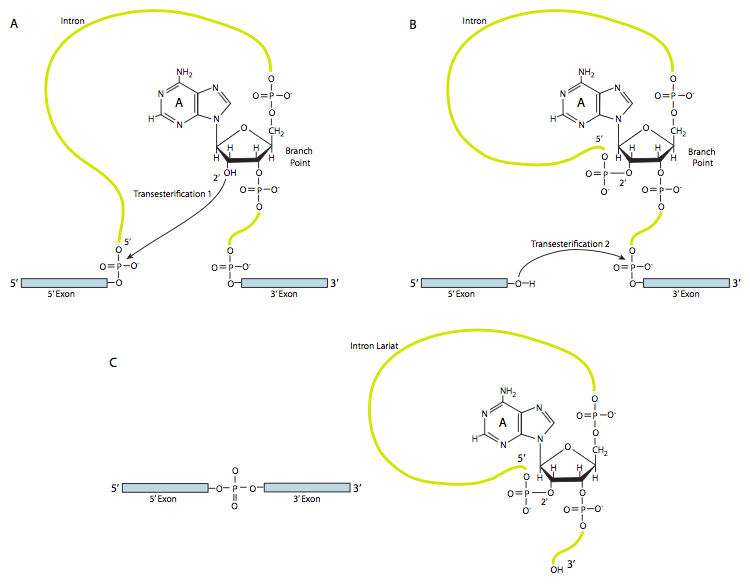
In the first step, the U1 snRNP binds to the intronic portion of the 5’ splice site. Next, the U2 snRNP binds to the consensus site around the branchpoint, but importantly, there is no base-pairing to the branchpoint A itself. Instead, due to basepairing of U2 with the surrounding sequence, the branchpoint A is forced to bulge out from the rest of the RNA in that region. U4, U5, and U6 join the spliceosome together, but while U5 binds to the 5’ exon, and U6 displaces U1 at the 5’ splice site, U4 is only transiently attached and also falls off the spliceosome before the first transesterification reaction. As the Figure shows, in this reaction, the 2’-OH of the branchpoint A nucleophilically attacks the 5’-phosphate of the first intron nucleotide to form a lariat structure in which the 5’ end of the intron is connected to the branchpoint via a 2’,5’-phosphodiester bond. This releases the 5’ exon (and the whole 5’ half of the RNA for that matter), but it is kept in close proximity to the 3’ exon (and the rest of the RNA) by U5, which attaches to both exons. This allows the second transesterification to take place, in which the 3’- OH of the first exon attacks the 5’ phosphate at the beginning of the second exon, thus simultaneously breaking the bond between the intron and the second exon, and also connecting the two exons via a conventional 3’,5’-phosphodiester bond. The intron, in the shape of a lariat, is thus released and will be quickly degraded.
Splicing is an efficient (with respect to genome size) way to generate protein diversity. In alternative splicing, some potential introns may be spliced out under certain circumstances but remain as coding sequence under other circumstances. Recall that the splice sites are recognized by base-pairing and therefore, there can be stronger and weaker splice sites depending on how close they are to the consensus and the complementary sequence on the snRNPs. Therefore, a gene with several potential introns may have all introns spliced out 80% of the time, but the other 20% of the time, perhaps only one or two introns are spliced out. Adding variability, there are splicing factors that may bind near splice sites and can either make them more easily recognizable, or nearly hidden.
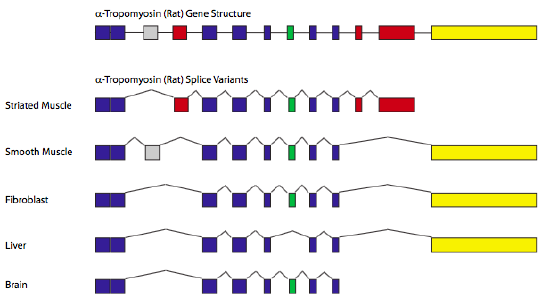
The classic example of alternative splicing is the gene encoding α-tropomyosin (Figure \(\PageIndex{11}\)). By splicing in/out different combinations of exons, a single gene can generate seven different proteins, depending on the tissue type. In these cases, particular types of cells or tissues contain specific combinations of splicing factors, and therefore control the recognition of specific splice sites, leading to the different splicing patterns.
Although this concludes the discussion of basic mechanisms of transcription, the next chapter is really a continuation of this one: control of gene expression in its simplest form is regulating the recognition of a promoter sequence by an RNA polymerase.