12.3: Actin Microfilaments
- Page ID
- 16165
Microfilaments are also known as actin filaments, filamentous actin, and f-actin, and they are the cytoskeletal opposites of the intermediate filaments. These strands are made up of small globular actin (g-actin) subunits that stack on one another with relatively small points of contact. You might envision two tennis balls, one fuzzy and the other covered in velcro hooks. Even if you push hard to mush them together, the area of contact between the balls (i.e. the area available for H-bonding between subunits) is fairly small compared to the overall surface area, or to the area of contact between IF subunits. They will hold together, but they can also fall apart with relatively little force. Contrast this with intermediate filaments, which might be represented as two ribbons of velcro hooks or loops. Considerably more work is required to take them apart. Because there are fewer H-bonds to break, the microfilaments can be deconstructed very quickly, making it suitable for highly dynamic applications.
When the actin subunits come together to form microfilaments, they interact directionally. That is, subunits have a “top” and a “bottom”, and the top of one subunit always interacts with the bottom of another. If we go to the “bottom”-most subunit of a filament, the open end is called the minus (-) end, while the opposite end, which incidentally sees more additive action, is called the plus (+) end. Microfilaments are also said to have polarity, but again this is only in the sense of having directionality, and has nothing to do with electrical charge. Individual microfilaments can exist, but most microfilaments in vivo are twisted pairs. Unlike DNA; however, microfilament pairs are not antiparallel: both strands have the same directionality.
The formation of filaments from g-actin is an ATP-dependent process, although not in the conventional sense of utilizing the energy released in hydrolysis. Instead, the globular actin subunits will only bind with another g-actin subunit if it has first bound an ATP. If the g-actin has bound ADP, then it must first exchange the ADP for ATP before it can be added onto a filament. This alters the conformation of the subunit to allow for a higher-affinity interaction. A short time later, hydrolysis of the ATP to ADP (with release of Pi) weakens the affinity but does not directly cause dissolution of the subunit binding. The hydrolysis is brought about by the actin itself, which has this ATPase enzymatic activity built in.
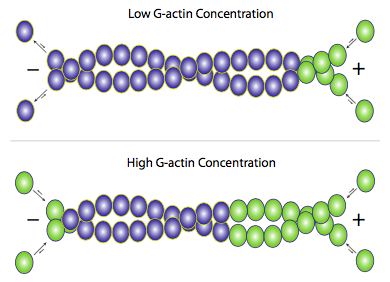
Although f-actin primarily exists as a pair of filaments twisted around each other, addition of new actin occurs by the addition of individual g-actin monomers to each filament (Figure \(\PageIndex{3}\)). Accessory proteins can be used to help or hinder either the building or breakdown of the filaments, but the primary mechanism is essentially self-regulating. When free g-actin levels are high, elongation of actin filaments is favored, and when the g-actin concentration falls, depolymerization of f-actin predominates. Under average physiological conditions, though, what is often seen in actin microfilaments is an effect called treadmilling. Since actin is mostly added onto one end but removed from the other, the net effect is that any given actin monomer in a filament is effectively moving from (+) end to (-) end even if the apparent length of the filament does not change.
In most cell types, the greatest concentration of actin-based cytoskeletal structures is found in the periphery of the cell rather than towards the center. This fits well with the tendency of the edges of the cell to be more dynamic, constantly adjusting to sense and react to its environment. Clearly, the polymerization and depolymerization of actin filaments is much faster than for intermediate filaments. The big exception to the actin-in-periphery rule is found in muscle cells. Actin filaments, and the myosin motor proteins that work on them, are the basis for muscle cell contraction, and ll up most of the muscle cells, not just the periphery. We will discuss the role of actin in both types of cell movement later in the chapter.


