11.3: Protein Folding in the Endoplasmic Reticulum
- Page ID
- 16158
The endoplasmic reticulum (ER) lumen plays four major protein processing roles:
- folding/refolding of the polypeptide,
- glycosylation of the protein,
- assembly of multi-subunit proteins, and
- packaging of proteins into vesicles.
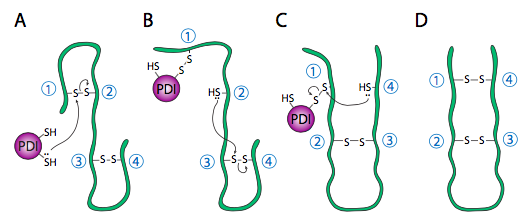
Refolding of proteins is an important process because the initial folding patterns as the polypeptide is still being translated and unfinished may not be the optimal folding pattern once the entire protein is available. This is true not just of H-bonds, but of the more permanent (i.e. covalent) disulfide bonds as well. Looking at the hypothetical example polypeptide, the secondary structure of the N-terminal half may lead to the formation of a stable disulfide bond between the first cysteine and the second cysteine, but in the context of the whole protein, a more stable disulfide bond might be formed between cysteine 1 and cysteine 4. The exchange of disulfide bonding targets is catalyzed by protein disulfide isomerase (PDI).
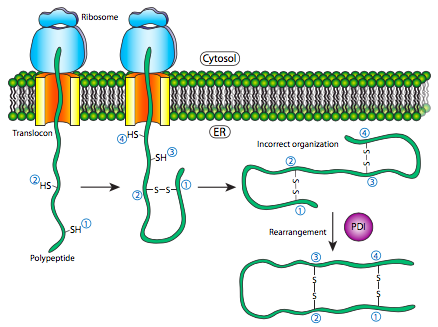
The internal redox environment of the endoplasmic reticulum, is significantly more oxidative than that in the cytoplasm. This is largely determined by glutathione, which is found in a 30:1 GSH:GSSG ratio or higher in the cytoplasm but at nearly 1:1 ratio in the ER lumen. This oxidative environment is also conducive to the disulfide remodeling. It should be noted that PDI does not choose the “correct” bonding partners. It simply moves the existing disulfide bonds to a more energetically stable arrangement. As the rest of the polypeptide continues to refold, breaking and making H-bonds quickly, new potential disulfide bond partners may move near one another and PDI can again attempt to rearrange the disulfide bonding pattern if the resulting pattern is more thermodynamically stable.
The assembly of multisubunit proteins and the refolding of polypeptides are similar in their use of chaperone proteins that help prevent premature folding, sequestering parts of the protein from H-bonding interaction until the full protein is in the ER lumen.
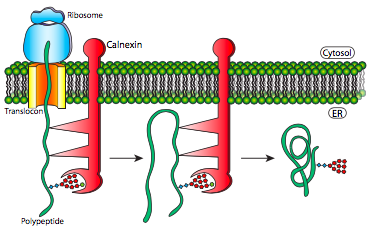
This mechanism simply makes finding the thermodynamically optimal conformation easier by preventing the formation of some potential suboptimal conformations. These chaperone proteins bind to the new proteins as they enter the lumen through the translocon and in addition to simply preventing incorrect bonds that would have to be broken, they also prevent premature interaction of multiple polypeptides with one another. This can be a problem because prior to the proper folding that would normally hide such domains within the protein, the immature polypeptides may have interaction domains exposed, leading to indiscriminate binding, and potentially precipitation of insoluble protein aggregates.
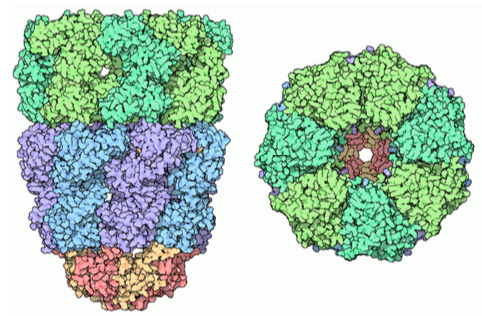
Chaperone proteins can also be found in prokaryotes, archaea, and in the cytoplasm of eukaryotes. These are somewhat similar to each other, and function somewhat differently than the types of folding proteins found in the ER lumen. They are referred to generally as chaperonins, and the best characterized is the GroEL/ GroES complex in E. coli. As the structure in Figure \(\PageIndex{11}\) indicates, it is similar in shape to the proteasome, although with a completely different function. GroEL is made up of two stacked rings, each composed of 7 subunits, with a large central cavity and a large area of hydrophobic residues at its opening. GroES is also composed of 7 subunits, and acts as a cap on one end of the GroEL. However, GroES only caps GroEL in the presence of ATP. Upon hydrolysis of the ATP, the chaperonins undergo major concerted conformational changes that impinge on the protein inside, causing refolding, and then the GroES dissociates and the protein is released back into the cytosol.