8.2: Prokaryotic Transcription
- Page ID
- 16136
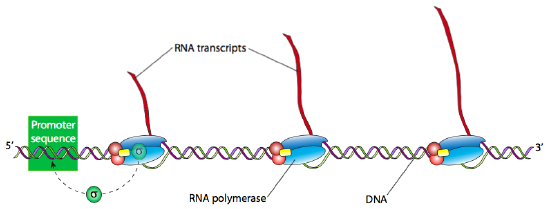
In E. coli, as with other prokaryotes, there is only one true RNA polymerase (not including the specialty RNA polymerase, primase, which makes short RNA primers for DNA replication). The polymerase is a multi-subunit holoenzyme comprised primarily of two α subunits, a β subunit, a β’ subunit, an ω subunit, and a σ subunit. The α subunits are primarily structural, assembling the holoenzyme and associated regulatory factors. The β subunit contains the polymerase activity that catalyzes the synthesis of RNA, while the β’ subunit is used to nonspecifically bind to DNA. The ω subunit is involved in assembly of the holoenzyme and may also play a role in maintaining the structural integrity of the RNA polymerase. Finally, there is the σ subunit, which does not stay closely associated with the core enzyme (αββ’ω) except when helping to initiate transcription, and is used to recognize the promoter by simultaneously decreasing the affinity of RNAP to DNA in general, but increasing the affinity of RNAP for specific DNA promoter sequences. Why decrease the affinity for non-specific DNA? When the RNAP is not in use, it does not just float about in the nucleoplasm: it is bound quite tightly along the DNA. When the sigma is bound, the decreased affinity allows the RNAP holoenzyme to move along the DNA and scan for promoter sequences. There are multiple isoforms of the σsubunit (such as the sigma-70 mentioned above), each of which recognizes different promoter sequences. All isoforms perform the same basic function of properly locating the RNAP to the start of a gene, and all isoforms only stay attached to the holoenzyme for that one transient purpose, after which they are released (usually after transcribing about ten nucleotides).
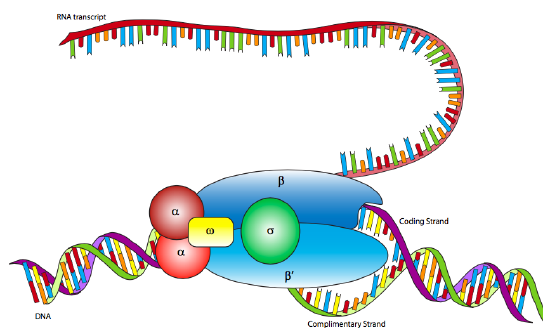
Although RNA polymerase was discovered in 1960, the E. coli RNAP has not yet been successfully mapped by x-ray crystallography. However, it is very similar to the RNAP of the archaean species, Thermophilus aquaticus, which is highly stable (= easier to crystallize) and for which an x-ray crystallographic structure has been elucidated. The data from the Taq RNAP structure and electron microscopic analyses of E. coli RNAP produce a lobster-claw-shaped holoenzyme. The inner surface of the claw is lined with positively charged amino acids that can interact with the negatively charged DNA, and when the holoenzyme binds a sigma subunit, the two halves of the claw (formed mostly by the beta and beta’ subunits) move closer together to interact with the DNA.
Rifamycins are a class of antibiotics that include rifamycin B, made by the bacteria Streptomyces mediterranei (incidentally just one of many antibiotics derived from the Streptomyces genus), and rifampicin, its synthetic cousin. They work by binding within the DNA-RNA channel near the active site of RNA polymerase, which sterically prevents the addition of nucleotides to the RNA strand. If the organism cannot transcribe RNA, it cannot use the RNA to make the enzymes and other proteins necessary for life either, and dies. The rifamycin binding site is highly conserved in most prokaryotes but not in eukaryotes, so the antibiotic kills bacteria specifically with little chance of harm to eukaryotes.
Once the holoenzyme has recognized and bound tightly to the DNA at the promoter site, the next step is to “melt” the DNA (breaking the H-bonds and separating the strands of the double helix) in that area so that the RNAP can proceed downstream, read the template DNA strand, and produce the new RNA. Often many RNA transcripts of a gene are needed to produce a large number of active proteins in a short span of time. Highly transcriptionally active genes therefore often have multiple RNA polymerases reading them, one right after another. Generally, an RNA polymerase only needs to process about 15 nucleotides before there is room for another RNAP can bind the promoter and start another transcript.
Strand separation is an energetically difficult process due to the strength of the combined H-bonds between the strands, and often an RNAP may make several short-lived abortive attempts before finally prying open the double helix long and far enough to allow the RNAP to stabilize and transcribe continuously to the stop site.
The elongation phase of transcription proceeds in a 5’ to 3’ direction, which is to say that new nucleotides are added to the 3’-OH of the growing strand. Elongation is a stochastic process in which one of the plentiful free- floating ribonucleotides drops into the active site of RNAP opposite the DNA template. If it is the correct nucleotide (complementary to the template), then H-bonds will temporarily form, stabilizing the new nucleotide in place long enough for the RNAP to catalyze the formation of a phosphodiester bond between the 3’-OH of the RNA-in-progress and the 5’-phosphate of the nucleotide. However, if it is the incorrect nucleotide, the proper H-bonds do not form, and the nucleotide usually dissociates from the active site before the RNAP has a chance to bind it to the growing RNA strand. Obviously, this is not a perfect system, and in fact, the error rate for transcription is quite high at approximately 1 in 10000 nucleotides. Fortunately, the cell generally churns out many copies of RNA from any given gene very quickly (approximately 80 nucleotides per second), most of which are either error-free or have errors that do not affect the function of the end-product protein. Furthermore, unlike DNA, in which errors of replication get carried along from one generation of cells to the next, RNA is not a storage medium, and its transient nature means that even mutations that severely impact the protein function only affect the few proteins translated from that one RNA, not the proteins generated from other RNAs made from the same template gene, much less subsequent generations. In other words, to misappropriate a phrase from the movie Meatballs, “It just doesn’t matter.”
Eventually, the RNA polymerase reaches the end of the gene and stops transcribing. The termination site is usually marked by a sequence of 4-10 adenine (A) residues on the template strand, and some have a palindromic G-C-rich region that forms a hairpin loop just upstream of the series of adenines. In the first case, it is thought that the resulting string of A-U base pairs is unstable and may lead to the RNAP and the new RNA strand falling off the template DNA while at the same time, the hairpin structure may cause the RNAP to stop or pause, and this can also lead to it dissociating from the DNA. Only about half of all transcription termination sites are marked in this way though, and the others have no significant hairpin loops or easily recognizable sequences other than a series of G-C-rich regions. In this type of termination site, the enzymatic co-factor, rho, is required for termination, and so this is known as rho-dependent termination. Rho is an RNA-binding protein with helicase activity, so it is postulated that it effects termination by forcing the RNA strand off of the DNA template.


