8.1: Introduction to Transcription
- Page ID
- 16135
To obtain the genetic information in a form that is easily read and then used to synthesize functioning proteins, the DNA must first be transcribed into RNA (ribonucleic acid). As we saw in chapter 1, RNA is extremely similar to DNA, using some of the same nitrogenous bases (adenine, guanine, cytosine) as well as one unique to RNA, uracil. Notice that uracil is very similar to thymine (chapter 7, Figure1), particularly in the placement and spacing of the hydrogen-bonding atoms. Since it is the hydrogen-bonding interaction of these bases (i.e. base-pairing of guanine to cytosine, adenine to thymine/uracil) that forms the basis of information transfer from original DNA to daughter cell DNA, it is logical to expect that the same kind of base-pairing mechanism is used to move the information from a storage state in the double-stranded nucleic acid (DNA) to a more useful/usable state in the form of a single-stranded nucleic acid (RNA).
In contrast to its cellular role as a transient and disposable carrier of genetic information, RNA is thought to have been the primary molecule responsible for making life possible on earth. It has long been postulated that it served dual roles as both a repository of genetic information and as a rudimentary enzyme to act upon that information. Unfortunately, prebiotic chemists have been stymied for decades in coming up with a reasonable synthetic pathway by which RNA could arise from the simple molecules of the earth’s primordial “soup”. The key problem was that ribose could be synthesized, though not particularly efficiently, and bases could be synthesized, but there was no way to connect them together. The chemistry would not allow a condensation reaction between the bases and sugars. In 2009, by leaving behind the conventional idea that ribonucleotides must have been synthesized from ribose and purines/pyrimidines, Powner, Gerland, and Sutherland (Nature 459:239-242, 2009) showed that in fact, ribonucleotides could be synthesized from the chemical conditions of a newly formed earth. Rather than attempt to make each “part” and put them together, Powner et al synthesized a molecule that contained parts of what would eventually be both the ribose and a pyrimidine, 2-aminooxazole. Through a series of reactions that utilized phosphate as a catalyst and scavenger, all of which were clearly plausible in the current model of primordial earth, both ribocytidine and ribouridine were created. Of course, this is only the beginning, since this does not extend directly to the formation of purine nucleotides, but it is a very significant step in prebiotic chemistry, and an excellent example of the virtues of “stepping outside of the box” sometimes.
The process of copying DNA into RNA is called transcription. In both prokaryotes and eukaryotes, transcription requires certain control elements (sequences of nucleotides within the DNA) to proceed properly. These elements are a promoter, a start site, and a stop site. The need for a recognizable point to begin and a point to end the process is fairly obvious. The promoter is somewhat different. The promoter controls the frequency of transcription. If you imagine the needs of a cell at any given time, clearly not all gene products are needed in the same quantity at the same time. There must be a way to control when or if transcription occurs, and at what speed.
The bare-bones version of the process goes something like this: (1) special docking proteins recognize the promoter sequence and bind to it, unzipping a small section around the “start” site; (2) RNA polymerase binds to those special proteins and to the little bit of single-stranded DNA that has just opened up; (3) a helicase enzyme (part of, or attached to the polymerase) unzips the DNA; (4) the RNA polymerase follows behind the helicase, “reading” the DNA sequence, taking ribonucleotides from the environment, matching them against the DNA template, and if they match, adding them to the previous ribonucleotide or RNA chain. This continues until the polymerase reaches the stop site, at which point, it detaches from the template DNA, also releasing the newly made RNA copy of that DNA. Of course, if that was all there was to it, there wouldn’t be entire journals dedicated to studying RNA, its transcription, and the control of that transcription.
The sequence of the promoter is directly related to its function. There may be promoters for housekeeping genes (needed constantly, but at low copy number), “normal” genes (needed as the cell’s situation dictates, rate of transcription also varies), stress response genes (needed rarely), and a variety of other categories. Even within a category, the sequence of the promoter determines its strength. This is based upon what is known as the “consensus sequence”. The consensus sequence is a theoretical “best” promoter based on a survey of all genes in a particular category. The Figure below shows an alignment of the promoter sequences of a variety of different genes, all of which are regulated by the same type of promoter and promoter-binding-protein. The highlighted boxes show areas centered around -35 (35 nucleotides upstream of the start site) and -10.
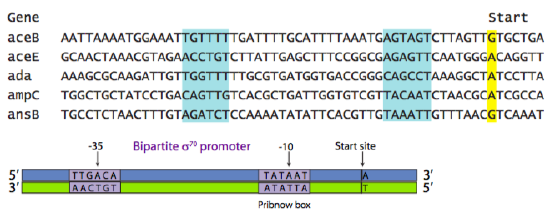
The consensus sequence in Figure \(\PageIndex{2}\) shows the most common nucleotide found at each position within those areas of similarity. In this example, the most common prokaryotic promoter is shown: the σ70 promoter, so called because it is recognized and bound by the σ70 transcription factor. [Here, and by universally accepted convention, recognition sequences of DNA are written as the nucleotides would occur from 5’ to 3’ on the sense, or non-template, strand.] It is a two-part promoter, with a region centered around -35 (consensus TTGACA), and a region (sometimes called Pribnow box, consensus TATAAT) centered around -10. The (-) sign indicates that the nucleotide is “upstream” of the start site. Upstream means “to the left” when the nucleotides are written as a string of letters, and it means “on the 5’ side of” with respect to the 5’-3’ directionality of a DNA strand. Notice the relationship between the various individual promoters and the consensus sequence. In general, those promoters with more matches to the consensus sequence are stronger promoters.
A few paragraphs ago, the task of the promoter was defined as controlling the frequency of transcription. How does it do that? What does it mean to be a stronger (or weaker) promoter? First, keep in mind that the expression of any given gene is not automatic, or 100%. At any point in time, many of a cell’s genes will be near 0%, or shut off. However, even genes that are turned on are transcribed at different rates. One of the governing factors is the recognition of the promoter site by the RNA Polymerase. For stronger promoters, the RNA polymerase is more likely to recognize the site, dock properly, open up the double helix, and begin transcribing. On the other hand, the RNA polymerase can potentially recognize weaker promoters, but it is less likely to do so, instead passing it by as just another unimportant stretch of DNA. While this is partially a matter of recognition by the polymerase, keep in mind that it is actually governed by recognition of the promoter sequence by the general transcription factors (to be discussed shortly) such as sigma factors in prokaryotes that are recognized by the polymerase.
Notice that there is a high proportion of (A)denines and (T)hymines in the σ70 promoter sequences. This is true for many promoters in both prokaryotic and eukaryotic genes. As you probably suspected, this is advantageous because there are only two H-bonds between A-T pairs (as opposed to 3 H-bonds between G-C pairs), which means that it is 33% easier to unzip.


