7.4: Eukaryotic Replication
- Page ID
- 16131
Given the crucial nature of chromosomal replication for life to exist, it is not surprising to find that eukaryotic DNA replication is very similar to the prokaryotic process. This section will highlight some of the differences, which are generally elaborations on the prokaryotic version.
Unlike prokaryotes, eukaryotic chromosomes often have multiple origins of replication. Considering the size of eukaryotic chromosomes, this is necessary to finish complete replication in a timely manner. Each of these origins defines a replicon, or the stretch of the DNA that is replicated from a particular origin. The replicons do not replicate at exactly the same time (although all within the same phase of the cell cycle, see chapter 15), so it is important to make sure that replicons are used only once during a cell cycle.
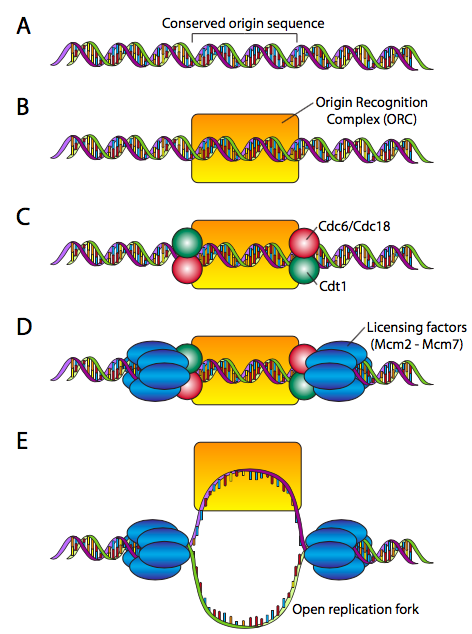
This requires a “licensing” mechanism. During the cell cycle phase before DNA replication is initiated, a pre-replicative complex is assembled at each origin (Figure \(\PageIndex{13}\)). The origins are highly variable in composition and length, ranging from ~100 to well over 10000 base pairs. The pre-RC proteins, on the other hand, are very highly conserved. The pre-RC begins by making the ORC (origin recognition complex, not a creature battling Frodo and Aragorn), which is comprised of six subunits (Orc1-Orc6). Although there is not significant sequence homology, the ORC approximates the function of the DnaA protein in E. coli. To complete the pre-RC, the ORC recruits a pair of proteins, Cdc6 and Cdt1 to each side, and they bind the MCM complex (a hexamer of Mcm2-Mcm6 that has an inactive helicase activity), leading to the fully licensed pre-RC. The origin is now ready for activation.
Activation of the pre-RC at an origin of replication requires first Mcm10, which facilitates protein kinases that phosphorylate the MCM complex, activating the helicase activity, and making the replication fork ready to accept the replication machine.
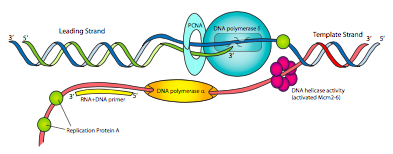
The components of the eukaryotic replication machine may have different names from the prokaryotic, but the functions should be very familiar (Figure \(\PageIndex{14}\)). There is a primase to make RNA primers, and tightly associated with the primase is DNA polymerase α. Pol α has neither a 5’-3’ exonuclease nor a 3’-5’ proofreading exonuclease, but it can synthesize DNA. However, this is not the primary DNA polymerase. The primase/pol a complex is essentially a fancy primase that begins with a short <10nt RNA primer and then adds another short ~15nt DNA fragment, making a hybrid primer.
Replication factor C (RFC), acting like the prokaryotic clamp loader complex, then replaces pol α with PCNA, the eukaryotic version of the β clamp. This then recruits DNA polymerase δ, which is the primary replicative DNA polymerase, equivalent to prokaryotic Pol III in function, and necessary for both leading and lagging strand synthesis. Finally, instead of SSB, eukaryotic cells use replication protein A (RPA) to organize and control single-stranded DNA as it is generated during the replicative process.
You may have noticed that none of the eukaryotic DNA polymerases discussed so far have a 5’ to 3’ exonuclease activity as used by prokaryotic DNA polymerase I to remove primers. In place of that, RNaseH1 and and FEN-1 remove the RNA primers (all but one ribonucleotide, and the last ribonucleotide, respectively). Interestingly, FEN-1 also excises chunks of lagging strand DNA within about 15 base pairs of the RNA if they contain mistakes. This seems to help alleviate the problem of lower fidelity of replication by pol α, which has no proofreading capability. After RNaseH1 and FEN-1 have removed primers and near-primer mistakes, pol δ fills in the missing nucleotides, and a ligase enzyme joins the fragments. Pol ε does have a 3’-5’ exonuclease activity that chops single-stranded DNA into small oligonucleotide fragments and is also associated with the replication machine. The function of Pol ε is not clearly understood.


