5.4: Oxidative Phosphorylation
- Page ID
- 16117
Oxidative phosphorylation denotes the phosphorylation of ADP into ATP, utilizing the energy from successive electron transports (hence the “oxidative”). The basic concept is that oxidation of NADH, being highly exergonic, can generate the energy needed to phosphorylate ADP. Since oxidation of NADH by oxygen can potentially release 52 kCal/mol (218 kJ/mol), and the energy needed to phosphorylate ATP is approximately 7.5 kCal/mol (30.5 kJ/mol), we should be able to expect the formation of several ATP per oxidized NADH. Indeed, this is what happens, although not directly. As noted with the breakdown of glucose, a one-step oxidation would generate too much energy for cellular processes to handle, and most would be wasted. So instead of oxidizing NADH directly with O2, the electrons are transferred to a series of gradually lower-energy carriers until finally reaching oxygen. This sequence is the electron transport chain.
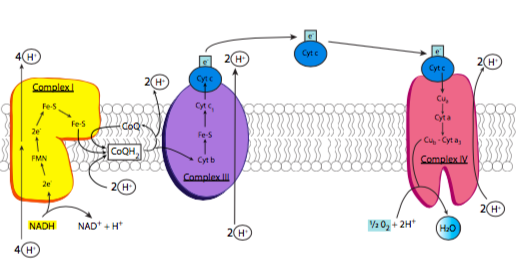
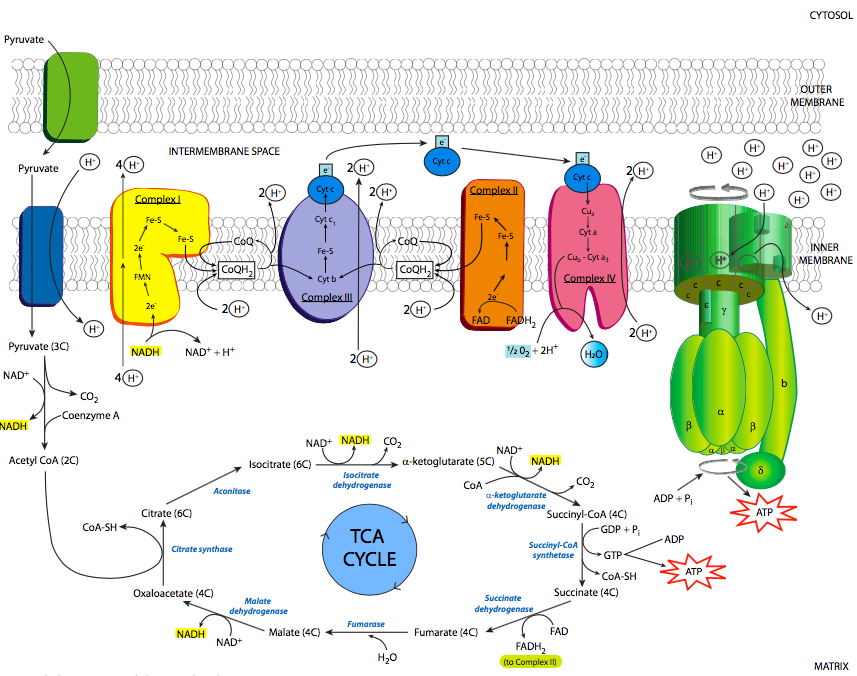
The electron transport chain is based on the activity of four major enzyme complexes (conveniently called complexes I-IV) embedded in the inner mitochondrial membrane, along with some small easily diffusible electron carriers to move the electrons from one complex to the next. These complexes are present in extremely high numbers as befits their necessity in generating energy, comprising nearly 75% of the inner membrane mass (in comparison, the plasma membrane of an average eukaryotic cell has a protein concentration closer to 50%). An overview of the process is shown in Figure \(\PageIndex{6}\): as previously noted, electrons are stripped from NADH, and eventually end up on oxygen. As the electrons are moved to lower-energy carriers, energy is released and used to pump protons from the mitochondrial matrix into the intermembrane space.
Complex I is an NADH dehydrogenase. Shown in yellow in Figure \(\PageIndex{6}\), its purpose is to remove a pair of electrons from NADH and transfer them onto ubiquinone (Coenzyme Q or CoQ), a small hydrophobic electron carrier that can then carry the electrons to complex III. This is a multistep process that involves first transferring the electrons onto an associated flavin mononucleotide (FMN) molecule, which then transfers the electrons to a set of iron-sulfur moieties connected to the enzyme complex itself (structure in Figure \(\PageIndex{7}\)). Finally, the electrons are moved onto ubiquinone. As these transfers occur, the energy that is released during these transfers powers the pumping of 4 H+ ions across the inner mitochondrial membrane. Complex I is inhibited by rotenone, a pesticide used primarily against insects and shes.
We’ll take a mental pass on complex II for now and hit it at the end of this roll call. The reasons will be apparent then.
Complex III is also known as the cytochrome bc1 complex (Figure \(\PageIndex{6}\), purple). The purpose of this complex is to pass the electrons from ubiquinone onto cytochrome c. The use of ubiquinone is important here, because it is stable with either two, or just one, extra electron. Cytochrome c, on the other hand, can only carry one electron. So, this complex docks ubiquinone, and holds it until it has passed its first electron onto cytochome c, which then moves onto complex IV, and then its second electron onto another cytochrome c. With each transfer, two protons are pumped across the membrane.
Finally, cytochrome c drops the electron off to complex IV, cytochrome c oxidase (Figure \(\PageIndex{6}\), red). Cytochrome c oxidase accomplishes the final step: transferring electrons onto oxygen atoms to make water. The really interesting thing about this process is that the enzyme must hold onto the electrons as they are transferred one at a time from cytochrome c, until it holds four electrons. Then, it can transfer one pair to each of the oxygen atoms in molecular oxygen (O2). It is very important to do this because transferring any less than all four electrons would lead to the creation of reactive oxygen species (ROS) that could cause damage to the enzymes and membranes of the mitochondria.
In fact, some well known poisons act at exactly this point. Both cyanide and carbon monoxide can bind with higher affinity than oxygen at the heme in complex IV. Since neither can accept electrons, the effect is just as though no oxygen was available.
Although cytochrome c oxidase is sometimes abbreviated COX, it is not the target of the COX-2 inhibitors that are used pharmaceutically in pain management, e.g. Bextra, Celebrex, or Vioxx. That refers to a family of enzymes known as the cyclooxygenases.
Oxygen is absolutely required. If oxygen is not available, there is no place to transfer the electrons, and very quickly, the electron transport chain is halted and carriers such as cytochrome c and CoQ cannot release their electrons and eventually there are no more available carriers. Similarly, when that happens, NAD+ is not regenerated, so the TCA cycle is also stuck. This leaves only the anaerobic non-oxygen-requiring glycolysis- fermentation cycle for generating ATP.
We now return to complex II (see Figure \(\PageIndex{10}\)). We mentioned complex II as succinate dehydrogenase when discussing the TCA cycle. It also participates in the electron transport chain by passing electrons to ubiquinone. However, rather than transferring electrons that originated from NADH like the other three complexes of the electron transport chain, the electrons originate from the covalently bound electron carrier FADH2 (flavin adenine dinucleotide), which received the electrons from succinate, as described in the TCA cycle section. Once the electrons have been passed to ubiquinone, it then moves on to complex III to drop off those electrons to cytochrome c, and the rest of the electron transport chain continues. FAD, the oxidized form of FADH2, is then ready to participate in the next redox cycle.
The purpose of this electron transport chain, with respect to ATP generation, is the pumping of H+ from the mitochondrial matrix into the intermembranous space. Since the concentration of protons is higher in the intermembrane space, it will take energy to move them against the concentration gradient, which is where our high-energy electrons come into the picture. As they move from one carrier to the next, they are moving from a higher to a lower energy state. This implies that some energy is lost from the electron, and some of that energy is tapped by the enzymes of the electron transport chain to move protons from the matrix to the intermembrane space.
There are two methods by which the protons are moved: the redox loop, and the proton pump. The proton pump, which is the method by which complex IV moves protons, is the easier to understand: H+ is bound on the matrix side of the enzyme in its reduced state (after it has received an electron), and a conformational shift occurs upon reoxidation to open the enzyme up to the intermembrane side, and the H+ is released. The redox loop, which occurs in complex I, and in complex III in a variation called the Q cycle, essentially posits that an initial redox center requires the binding of both the high energy electron and a proton from the matrix side of the membrane. When the electron is transferred to the next redox center in the chain, a proton is released to the intermembrane space.
Whatever the mechanism, what is the point of all this proton pumping? As you might suspect, using up energy to pump an ion against its concentration gradient isn’t done for the fun of it. Rather, this generates significant potential energy across the inner mitochondrial membrane. And, it so happens that there is an enzyme that can convert that energy into the physiologically useful chemical form of ATP. This enzyme is, not surprisingly, named ATP synthase (Figure \(\PageIndex{8}\)). It is also referred to in some texts as the F1F0-ATPase, based on its reverse activity (at the expense of ATP, it can pump protons), and the fact that it can be broken down into two major functional units: F1 which can hydrolyze but not synthesize ATP and is a soluble protein, and F0 which is an insoluble transmembrane protein.
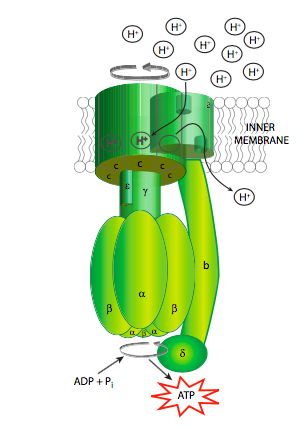
The ATP synthase is an extraordinary example of an enzyme that transforms the energy inherent in a concentration gradient across a membrane into mechanical energy, and finally into chemical bond energy. It is descriptively called a “rotary engine” because the very generalized sequence of events is as follows: protons ow down their gradient through a proton channel subunit of the ATP synthase, in owing down the gradient, energy is released, this energy causes rotation of a multisubunit “wheel”-like subunit attached to a spindle/axle (g subunit) which also spins. The spinning of this asymmetrically shaped spindle unit causes conformational changes in the catalytic subunit (made of the a and b subunits) it is attached to, changing an ADP+Pi binding site to a catalytic site that can “squeeze” the molecules together into an ATP, and then finally open up to release the ATP (Figure \(\PageIndex{9}\)).
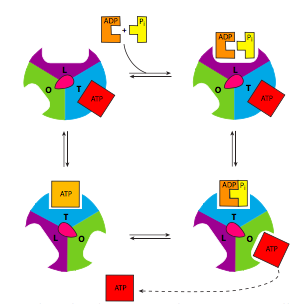
Of course, it isn’t quite that simple (Figure \(\PageIndex{8}\)). Starting with the initial movement of pro- tons, as they move from the intermembrane space into the ATP synthase, they enter a small hydrophilic channel (a) and then bind onto one of the c-subunits of the “water wheel” c-ring. Binding of the H+ to the c-subunit causes it to lose affinity for the a- subunit, allowing it to spin, and simultaneously causes a conformational change that essentially pushes off against the a-subunit, initiating the movement. Once it has spun around almost a complete turn, the H+ is positioned by another channel (b), which funnels it from the c-subunit into the matrix. The c-subunit structure is connected to an asymmetric spindle that is itself connected to the catalytic subunits.


