11.2: Protein Trafficking
- Page ID
- 16157
The idea that propeptide sequences have important functions in protein maturation beyond just keeping them from being active is not exclusive to assembly. A major class of cleaved peptide sequences is signal peptides. Signal peptides direct the protein from the cytoplasm into a particular cellular compartment. In the case of prokaryotes, this essentially means the cell membrane, but for eukaryotes, there are specific signal peptides that can direct the protein to the nucleus, to the mitochondria, to the endoplasmic reticulum, and other intracellular organelles. The peptides are specifically recognized by receptors on the membranes of particular compartments, which then help to guide the insertion of the protein into or through the membrane. Almost all protein synthesis in eukaryotes is carried out in the cytoplasm (with the exception of a few proteins in the chloroplasts and mitochondria), so proteins found in any other compartment or embedded in any membrane must have been targeted and transported into that compartment by its signal sequence.
Although this is primarily considered a eukaryotic process given that there are so many potential targets, prokaryotes do have membrane proteins (in fact, some 800 different ones in E. coli comprising ~20% of total protein), and they are positioned there with the aid of insertase enzymes such as YidC and complexes such as Sec translocase. The Sec translocase uses a signal recognition particle (SRP) much like that in eukaryotes, and will be discussed later in this chapter when the SRP is introduced. YidC, which has eukaryotic homologues (e.g. Oxa1 in mitochondria), is a 61 kDa transmembrane protein that is placed in the membrane through an SRP-Sec translocase mechanism. Once there, YidC interacts with nascent polypeptides (once they reach ~70 amino acids long) that have begun to interact with the lipids of the cell membrane, and pushes the protein into/through the membrane.
The nucleus is one such compartment, and examples of the proteins found within include DNA and RNA polymerases, transcription factors, and histones. These and other nuclear proteins have an N-terminal signal sequence known as the NLS, or nuclear localization signal. This is a well-studied pathway that involves a set of importin adapter proteins and the nuclear pore complex (Figure \(\PageIndex{3}\)). Transport into the nucleus is particularly challenging because it has a double membrane (remember that it is contiguous with the endoplasmic reticulum membrane. Although there are other mechanisms for making proteins that are embedded in the nuclear membrane, the primary mechanism for import and export of large molecules into and out of the nucleus itself is the nuclear pore complex. The complex is very large and can be made of over 50 different proteins (nucleoporins, sometimes called nups). The nucleoporins are assembled into a large open octagonal pore through the nuclear membranes. As Figure \(\PageIndex{3}\) indicates, there are antenna-like fibrils on the cytoplasmic face, and these help to guide proteins from their origin in the cytoplasm to the nuclear pore, and on the nuclear side there is a basket structure. Of course, not all proteins are allowed into the nucleus, and the mechanism for distinguishing appropriate targets is straightforward. The protein must bear a nuclear localization signal (NLS). While in the cytoplasm, an importin-α protein binds to the NLS of a nuclear protein, and also binds to an importin-β. The importin-β is recognized and bound by the nuclear pore complex. The details of the transport mechanism are murky, but phenylalanine-glycine repeats in the nucleoporin subunits (FG-nups) are thought to be involved.
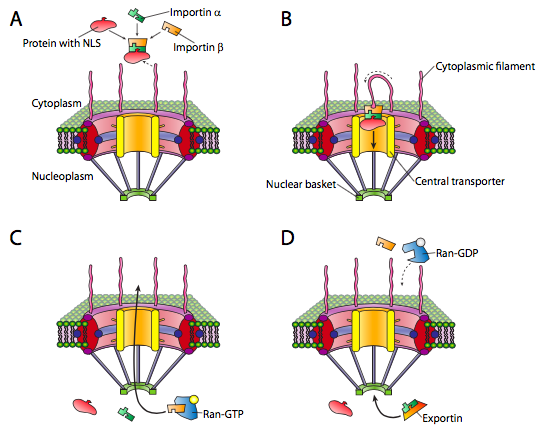
Once the nucleoprotein-importin aggregate is moved into the nucleus, Ran-GTP, a small GTPase, causes the aggregate to dissociate (Figure \(\PageIndex{3}\)c). The imported protein is released in the nucleus. The importins are also released in the nucleus, but they are exported back out again to be reused with another protein targeted for the nucleus.
The mechanisms of small GTPase activation of other processes will be discussed again in more detail in later chapters (cytoskel- eton, signaling). The key to understanding the mechanism is to remember that the GTPase hydrolyzes GTP to GDP, but still holds onto the GDP. Although the GTPase will hydrolyze GTP spontaneously, the GTPase-activating protein, GAP (or Ran-GAP in this case) greatly speeds the rate of hydrolysis. In order to cycle the system back to GTP, the GDP is not re-phosphorylated: it is exchanged for a new GTP. The exchange is greatly facilitated by the action of an accessory protein, the guanine nucleotide exchange factor (GEF), in this particular case, a Ran-GEF.
Export from the nucleus to the cytoplasm also occurs through the nuclear pore. The Ran-GTP is also a part of the export complex (Figure \(\PageIndex{3}\)d), and in conjunction with an exportin protein and whatever is to be exported, is moved out of the nucleus via the nuclear pore. Once in the cytoplasm, the hydrolysis of GTP to GDP by Ran (activated by Ran-GAP, a cytoplasmic protein) provides the energy to dissociate the cargo (e.g. mRNA) from the exporting transport molecules. The Ran-GDP then binds to importins, re-enters the nucleus, and the GDP is exchanged for GTP.
The nuclear pore is the only transport complex that spans dual membrane layers, although there are coordinated pairs of transport complexes in double-membraned organelles such as mitochondria. The transport proteins in the outer mitochondrial membrane link with transport proteins in the inner mitochondrial membrane to move matrix-bound proteins (e.g. those involved in the TCA cycle) in from the cytoplasm. The complexes that move proteins across the outer membrane are made up of Tom (translocator outer membrane) family of proteins. Some of the proteins will stay embedded in the outer membrane: they are processed by a SAM (sorting and assembly machinery) complex also embedded in the outer membrane). Meanwhile, others continue to the Tim (translocator inner membrane) proteins that move them across the inner membrane. As with the nuclear proteins, there is a consensus signal sequence on mitochondrial proteins that is bound by cytosolic chaperones that bring them to the Tom transporters. As shown in the table below, there are signal sequences/propeptides that target proteins to several other compartments.
Of particular importance for the rest of this chapter, is the sequence targeting proteins to the endoplasmic reticulum, and by extension, any proteins destined for the ER, the Golgi apparatus, the cell membrane, vesicles and vesicularly-derived compartments, and secretion out of the cell. Here, in addition to an N-terminal signal sequence, the position of secondary internal signal sequences (sometimes called signal patches) helps to determine the disposition of the protein as it enters the ER.
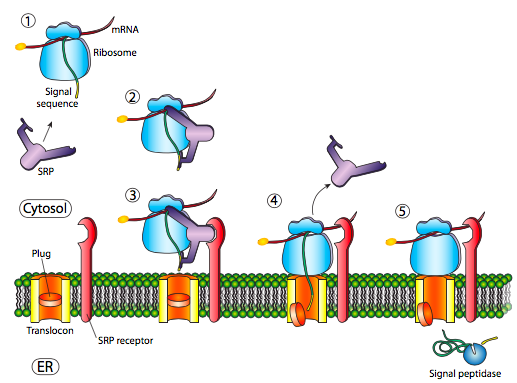
The initial insertion requires recognition of the signal sequence by SRP, the signal recognition protein. The SRP is a G-protein and exchanges its bound GDP for a GTP upon binding to a protein’s signal sequence. The SRP with its attached protein then docks to a receptor (called the SRP receptor, astoundingly enough) embedded in the ER membrane and extending into the cytoplasm. The SRP usually binds as soon as the signal sequence is available, and when it does so, it arrests translation until it is docked to the ER membrane. Incidentally, this is the origin of the “rough” endoplasmic reticulum: the ribosomes studding the ER are attached to the ER cytoplasmic surface by the nascent polypeptide it is producing and an SRP. The SRP receptor can exist on its own or in association with a translocon, which is a bipartite translocation channel. The SRP receptor (SR) is also a GTPase, and is usually carrying a GDP molecule when unassociated. However, upon association with the translocon it exchanges its GDP for a GTP. These GTPs are important because when the SRP binds to the SR, both GTPase activities are activated and the resulting release of energy dissociates both from the translocon and the nascent polypeptide. This relieves the block on translation imposed by the SRP, and the new protein is pushed on through the translocon as it is being synthesized. Once the signal sequence has completely entered the lumen of the ER, it reveals a recognition site for signal peptidase, a hydrolytic enzyme that resides in the ER lumen and whose purpose is to snip off the signal peptide.
Prokaryotes also use an SRP homolog. In E. coli, the SRP is simple, made up of one protein subunit (Ffh) and a small 4.5S RNA. By comparison, some higher eukaryotes have an SRP comprised of six different proteins subunits and a 7S RNA. Similarly, there is a simple prokaryotic homologue to the SRP receptor, FtsY. An interesting difference is that FtsY generally does not interact with exported proteins, and appears to be necessary only for membrane-embedded proteins. Otherwise, there are many similarities in mechanism for SRP-based insertion of membrane proteins in eukaryotic and prokaryotic species, including GTP dependence, and completion of the mechanism by a translocase (SecYEG in E. coli).
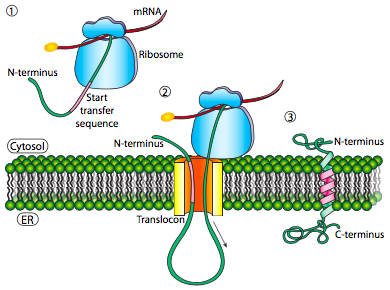
If that was the only signal sequence in the protein, the remainder of the protein is synthesized and pushed through the translocon and a soluble protein is deposited in the ER lumen, as shown in Figure \(\PageIndex{4}\). What about proteins that are embedded in a membrane? Transmembrane proteins have internal signal sequences (sometimes called signal patches). Depending on their relative locations, they may be considered either start-transfer or stop-transfer sequence, where “transfer” refers to translocation of the peptide through the translocon. This is easiest to understand by referring to Figure \(\PageIndex{5}\).
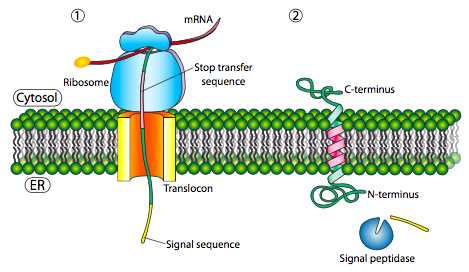
If there is a significant stretch of mostly-uninterrupted hydrophobic residues, it would be considered a stop-transfer signal, as that part of the protein can get stuck in the translocon (and subsequently the ER membrane) forcing the remainder of the protein to remain outside the ER. This would generate a protein that inserts into the membrane once, with its N-terminus in the ER lumen and the C-terminus in the cytoplasm. In a multi-pass transmembrane protein, there could be several start- and stop- transfer hydrophobic signal patches.
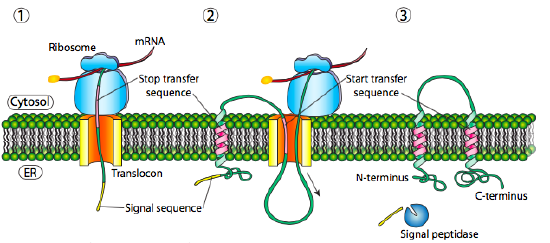
Building on the single-pass example, if there was another signal patch after the stop-transfer sequence, it would act as a start-transfer sequence, attaching to a translocon and allowing the remainder of the protein to be moved into the ER. This results in a protein with both N- and C- termini in the ER lumen, passing through the ER membrane twice, and with a cytoplasmic loop sticking out. Of course, the N-terminus could be on the other side. For a cytoplasmic N-terminus, the protein cannot have an N-terminal signal sequence (Figure \(\PageIndex{7}\)). It has an internal signal patch instead. It plays essentially the same role, but the orientation of the patch means that the N-terminal stays cytoplasmic. The polypeptide translated after the patch is fed into the ER. And just as in the last example, multiple stop- and start- sequences can reinsert the protein in the membrane and change the facing of the next portion.