8.3: Eukaryotic Transcription
- Page ID
- 16137
Transcription in eukaryotes is more complicated, but follows the same general ideas. The promoter sequences are much more varied both in placement (with respect to the start site) and size. As we will see in the next chapter, eukaryotic genes have many more control elements regulating their expression than do prokaryotic genes. Not only are there more control elements, there are also more RNA polymerases, which serve different specific cellular functions. Obviously, the broad function and location of all the RNA polymerases is the same: read a DNA template and transcribe an RNA copy of it; and since the DNA is found only in the nucleus, so are the polymerases. However, the polymerases differ in exactly what kinds of RNA they produce. RNA Poly- merase I is specialized for producing pre-rRNA (rRNA = ribosomal RNA). The pre-rRNA is cleaved post-transcriptionally and incorporated into the ribosomes. Since ribosomes are assembled in the the nucleolus, that is the part of the nucleus in which most RNA Polymerase I is concentrated. RNA Polymerase III also makes an RNA (5S) that is incorporated into the ribosome. It also makes other untranslated RNAs such as tRNAs and a variety of small nuclear RNAs. The only RNA polymerase that makes the translatable RNA (mRNA, or messenger RNA) that most people think of when RNA is referred to generically, is RNA polymerase II. This is the RNA polymerase that produces pre-mRNA, which after some processing, becomes mRNA, is transported out of the nucleus, and finally translated into proteins. All of the eukaryotic RNA polymerases are composed of two large subunits, roughly analogous to the β and β’ subunits of prokaryotic RNAP, but instead of just three or four other subunits, there are over a dozen smaller subunits to the eukaryotic RNA polymerase holoenzymes.
Initiation of transcription is also much more complicated. Not only is there great variety in promoters recognized by RNAP II, both RNAP I and RNAP III recognize promoters with particular structural characteristics.
The eukaryotic RNA polymerases were named I, II, and III based on their elution order from ion-exchange chromatography purification. They are also partially distinguishable by their sensitivity to α-amanitin and related amatoxin-family mushroom poisons. RNAP I (and prokaryotic RNAP) is insensitive to these toxins, RNAP III is somewhat sensitive (Kd ~10-6 M), and RNAP II is highly sensitive (Kd ~10-8 M). These toxins act by binding to a site in the RNA-DNA cleft and interfering with translocation of the RNA. That is, there is no problem with importing a nucleotide or with attaching it to the new RNA, but the RNA strand cannot move through the active site and allow the next nucleotide to be added.
One of the most common eukaryotic RNAP II promoters is the TATA box, named for the highly conserved motif that defines it. Although it appears similar to the Pribnow box in prokaryotes, it is generally located further upstream from the start site, and its position is far more variable. Whereas the Pribnow box is located at -10, the TATA box may be located closer to -30 +/- 4. Also, rather than just a sigma factor to recognize the promoter in conjunction with the polymerase core enzyme, the eukaryotic promoter is recognized by a multi-subunit complex called transcription factor IID (TFIID). TFIID is comprised of TATA-binding protein (TBP) and several TBP-associated factors (TAFs).
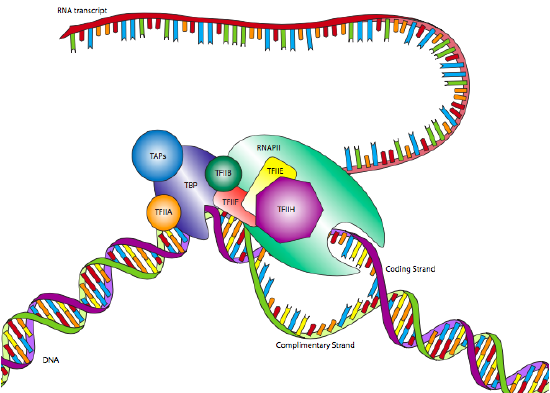
This binding of the promoter by TFIID occurs independently of RNA Polymerase II, and in fact, RNAP II will not attach to TFIID at this time. After TFIID has bound the TATA box, two more transcription factors, TFIIA and TFIIB, attach to the TFIID as well as the nearby DNA, stabilizing the complex. TFIIF attaches to TFIID and TFIIB to allow docking of the RNA Polymerase II. The complex is still not ready to begin transcription: two more factors are required. TFIIE binds TFIIF and RNAP II, and finally, TFIIH attaches to RNAP II, providing a helicase activity needed to pry apart the two strands of DNA and allow the polymerase to read one of them. TFIIH also has another important enzymatic activity: it is also a serine kinase that phosphorylates the carboxyl-terminal domain (CTD) of RNA polymerase II. There are several serines in the CTD, and as they are sequentially phosphorylated, the CTD extends like a (negatively charged) tail and helps to promote separation between the RNAP II and the TFIID/promoter.
Elongation of the RNA strand in eukaryotes is very similar to that in prokaryotes with the obvious difference that transcription occurs in the nucleus rather than in the cytoplasm. Thus, in prokaryotes, the RNA can be used for translation of proteins even as it is still being transcribed from the DNA! In eukaryotes, the situation is significantly more complex: there are a number of post-transcriptional events (5’ end-capping, 3’ polyadenylation, and often RNA splicing) that must occur before the RNA is ready to be transported out of the nucleus and made available for translation in the cytoplasm.
Termination of eukaryotic transcription is not well-described at this writing. RNAP I appears to require a DNA-binding termination factor, which is not analogous to the prokaryotic Rho factor, which is an RNA binding protein. RNAP III terminates transcription without any external factor, and this termination usually occurs after adding a series of uridine residues. However, it does not appear to use the hairpin loop structure found in rho-independent bacterial transcription. The termination of protein-coding RNAP II transcripts is linked to an enzyme complex that also cleaves part of the 3’ end of the RNA off, and adds a poly-A tail. However, it is not clear how the polyadenylation complex is involved in determining the point of transcription termination, which can be over 1000 nucleotides beyond the poly-A site (e.g. the β-globin gene in Mus musculus). Upon termination and release from the RNAP II and template DNA, the RNA is known as the primary transcript, but must undergo post-transcriptional processing before it is a mature messenger RNA (mRNA) ready to be exported to the cytoplasm and used to direct translation.


