1.4: Tour of the Eukaryotic Cell
- Page ID
- 16411
A. The Nucleus
The nucleus separates the genetic blueprint, i.e., DNA from the cell cytoplasm. Although the eukaryotic nucleus breaks down during mitosis and meiosis as chromosomes form and cells divide, it spends most of its time in interphase, the time between cell divisions. This is where the status of genes (and therefore of the proteins produced in the cell) is regulated. rRNA, tRNA and mRNA are transcribed from genes, processed in the nucleus, and exported to the cytoplasm through nuclear pores. Some other RNAs remain in the nucleus, typically participating in the regulation of gene activity. In all organisms, dividing cells must produce and partition copies of their duplicated genetic material equally between new daughter cells. Let’s look first at the structural organization of the nucleus, and then at its role in the genetics of the cell and of the whole organism.
1. Structure of the Interphase Nucleus
The nucleus is the largest organelle in the cell. A typical electron microscope image of a nucleus, the largest eukaryotic organelle in a cell, is shown below.
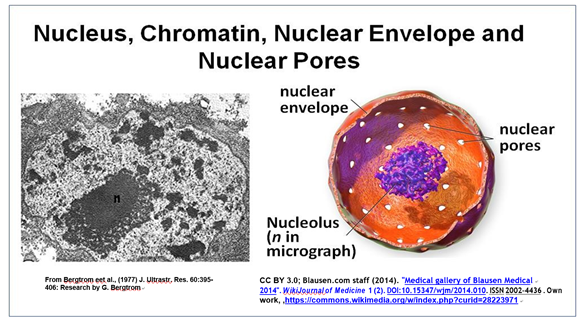
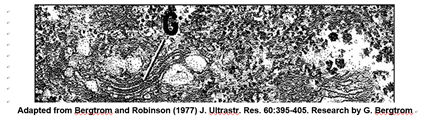
This cross-section of an interphase nucleus reveals its double membrane, or nuclear envelope. The outer membrane of the nuclear envelope is continuous with the RER (rough endoplasmic reticulum). Thus, the lumen of the RER iscontinuous with the space separating the nuclear envelope membranes. The electron micrograph also shows a prominent nucleolus (labeled n) and a darkly granular RER surrounding the nucleus. Zoom in on the micrograph; you may see the double membrane of the nuclear envelope. You can also make out ribosomes (small granules) bound to both the RER and the outer nuclear membrane. Nuclear envelope pores (illustrated in the cartoon at the right) allow large molecules and even particles to move in and out of the nucleus across both membranes.
The nucleus is not an unorganized space surrounded by the nuclear envelope, as seems to appear in the transmission electron micrographs. The nucleolus is just the largest of several nuclear inclusions that seem to segregate nuclear functions.
Santiago Ramón y Cajal reported more structures in the nuclei of neurons more than 100 years ago, drawing his observations before modern photomicrographic technology became widely available. See what he saw at Cajal's Nuclear Bodies, including the nucleolus and what came to be known as Cajal bodies (CBs). As we saw earlier, Ramón y Cajal shared the Nobel Prize in Physiology or Medicine 1906 with Camillo Golgi for their studies of nerve cell structure. Check out a gallery of Cajal’s hand-drawn micrographs of brain nerve cells in Cajal's Beautiful Brain Cells.
Later seen in an electron microscope, CBs look like coils of tangled thread, and were thus called coiled bodies (conveniently, also CBs). Other nuclear bodies since identified include Gems, PML bodies, nuclear speckles (or splicing speckles), histone locus bodies (HLBs) …, and more! Different nuclear bodies turn out to be associated with specific proteins. The localization of specific proteins to different nuclear bodies can be seen in the immunofluorescence micrograph below.
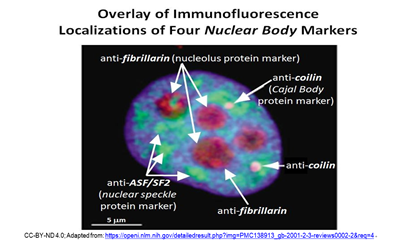
Nucleoli contain fibrillarin proteins and stain red because they have been treated with red-fluorescence-tagged antifibrillarin antibodies. CBs contain the protein coilin. They fluoresce pink because the nuclei were treated with fluorescence- tagged anticoilin antibodies. Green-fluorescent antibodies to the ASF/SF2 protein localize to nuclear speckles. As part of, or included in a nuclear matrix, nuclear bodies organize and regulate different aspects of nuclear activity and molecular function. The different nuclear bodies perform specific functions and interact with each other and with proteins DNA and RNA to do so. We will revisit some nuclear bodies in their working context in later chapters.
2. Every Cell (i.e., Every Nucleus) of an Organism Contains the Same Genes
We read earlier that bacteria are busy doubling and partitioning their naked DNA chromosomes at the same time as they grow and divide by binary fission. In eukaryotic cells, a cell cycle divides life into discrete consecutive events. During most of the cell cycle, cells are in interphase and DNA is wrapped up in proteins in a structure called chromatin. It is not merely the DNA, but chromatin that must be duplicated when cells reproduce. Duplication of DNA involves rearranging chromatin proteins. This occurs before cell division (mitosis and cytokinesis). As the time of cell division nears, chromatin associates with even more proteins, condensing to form chromosomes, while the nuclear envelope dissolves.
You may recall that every somatic cell of an organism contains paired homologous chromosomes, and therefore two copies of every gene an organism owns. On the other hand, sperm and eggs contain one of each pair of chromosomes, and thus one copy of each gene. Whether by mitosis or meiosis, cytokinesis separates duplicated chromosomes to daughter cells. In the fluorescence micrograph of a cell in the metaphase stage of mitosis (below), the chromosomes (blue) are just about to be pulled apart by microtubules of the spindle apparatus (green).
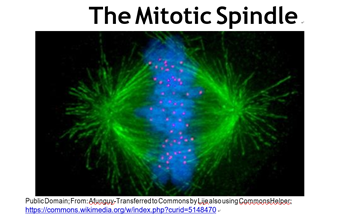
As the chromosomes separate and daughter cells form, nuclei reappear and chromosomes de-condense. These events mark the major visible difference between cell division in bacteria and eukaryotes. Cytokinesis begins near the end of mitosis. Sexual reproduction, a key characteristic of eukaryotes, involves meiosis rather than mitosis. The mechanism of meiosis, the division of germ cells leading to production of sperm and eggs, is similar to mitosis except that the ultimate daughter cells have just one each of the parental chromosomes, eventually to become the gametes (eggs or sperm).
A key take-home message here is that every cell in a multicellular organism, whether egg, sperm or somatic, contains the same genome (genes) in its nucleus. This was understood since mitosis and meiosis were first described in the late 19th century. However, it was finally demonstrated in 1962, when John Gurdon and Shinya Yamanaka transplanted nuclei from the intestinal cells the frog Xenopus laevis into enucleated eggs (eggs from which its own nucleus had been removed). These ‘eggs’ grew and developed into normal tadpoles, proving that no genes are lost during development, but just expressed differentially.
We will revisit animal cloning later in this book. But for now, it’s sufficient to know that Molly the cloned frog was followed in 1996 by Dolly, the first cloned sheep, and then other animals, all cloned from enucleated eggs transplanted with differentiated cell nuclei. Click Cloning Cuarteterra for the 60 Minutes story of the cloning of Cuarteterra, a champion polo mare whose clones are also champions! For their first animal cloning experiments, Gurdon and Yamanaka shared the 2012 Nobel Prize form Physiology or medicine.
B. Ribosomes
On the other end of the size spectrum, ribosomes are evolutionarily conserved protein synthesizing machines in all cells. They consist of a large and a small subunit, each made up of multiple proteins and one or more molecules of ribosomal RNA (rRNA). Ribosomes bind to messenger RNA (mRNA) molecules, moving along the mRNA as they translate 3-base code words (codons) to link amino acids into polypeptides. Multiple ribosomes can move along the same mRNA, becoming a polyribosome, simultaneously translating the same polypeptide encoded by the mRNA. The granular appearance of cytoplasm in electron micrographs is largely due to the ubiquitous distribution of ribosomal subunits and polysomes in cells.
The illustration below shows a ‘string’ of ribosomes, the polyribosome or polysome for short.
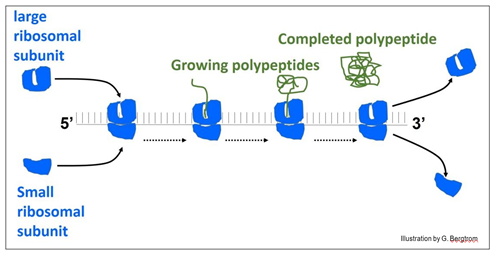
In the illustration, ribosomes assemble at the left of the messenger RNA (mRNA) to form the polysome. When they reach the end of the message, the ribosomes disassemble from the RNA and release the finished polypeptide.
In an electron micrograph of leaf cells from a quiescent desiccated dessert plant, Selaginella lepidophylla, you can make out randomly distributed ribosomes and ribosomal subunits (arrows, below left). In cells from a fully hydrated plant, you can see polysomes as more organized strings of ribosomes (arrows, below right).

Eukaryotic and prokaryotic ribosomes differ in the number of RNA and proteins in their large and small subunits, and thus in their overall size. Isolated ribosomes centrifuged in a sucrose density gradient move at a rate based on their size (or more specifically, their mass).
The illustration below shows the difference in ribosomal ‘size’, their protein composition and the number and sizes of their ribosomal RNAs.
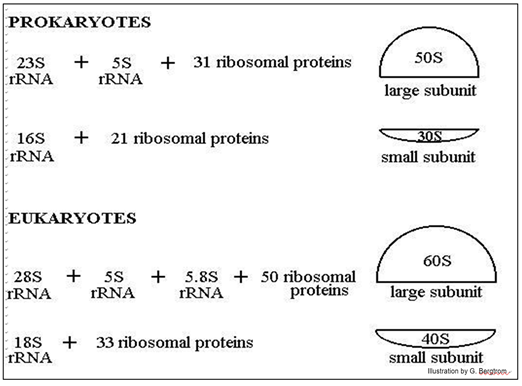
The position of ribosomal subunits in the gradient is represented by an S value, after Svedborg, who first used sucrose density gradients to separate macromolecules and particles by mass. Note that the ribosomal RNAs themselves also separate on sucrose density gradients by size, hence their different S values.
C. Internal membranes and the Endomembrane System
Microscopists of the 19th century saw many of these structures using the art of histology, staining cells to increase the visual contrast between cell parts. One of these, Camillo Golgi, an early neurobiologist, developed a silver (black) stain that first detected a network of vesicles we now call Golgi bodies (Golgi vesicles) in nerve cells. For their discoveries in cellular neuroscience, Golgi and Santiago Ramón y Cajal shared the 1906 Nobel prize for Medicine or Physiology.
Many vesicles and vacuoles in cells, including Golgi vesicles, are part of the endomembrane system. Proteins synthesized on the ribosomes of the RER (rough endoplasmic reticulum) can enter the interior space (lumen) or can become part of the RER membrane itself. Production of RER, SER (smooth endoplasmic reticulum), Golgi bodies, lysosomes, microbodies and other vesicular membranes, as well as their protein content all begin in the RER. The RER and protein contents bud into transport vesicles that fuse with Golgi Vesicles (G in the electron micrograph below).
In their journey through the endomembrane system, packaged proteins undergo stepwise modifications (maturation) before becoming biologically active (below).
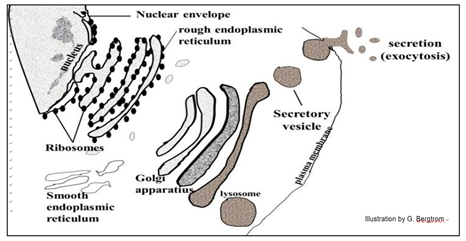
102 Golgi Vesicles & the Endomembrane System
Some proteins made in the endomembrane system are secreted by exocytosis. Others end up in organelles like lysosomes that contain hydrolytic enzymes. These enzymes are activated when the lysosomes fuse with other organelles destined for degradation. Food vacuoles form when a plasma membrane invaginates, engulfing food particles. They then fuse with lysosomes to digest the engulfed nutrients.
Autophagosomes are small vesicles that surround and eventually encapsulate tired organelles (for example, worn out mitochondria), eventually merging with lysosomes whose enzymes degrade their contents. In 2016, Yoshinori Ohsumi earned the Nobel Prize in Physiology and Medicine for nearly 30 years of research unraveling the cell and molecular biology of autophagy. Microbodies are a class of vesicles smaller than lysosomes, but formed by a similar process. Among them are peroxisomes that break down toxic peroxides formed as a by-product of cellular biochemistry. Some vesicles emerging from the RER will become part of the SER, which has several different functions (e.g., alcohol detoxification in liver cells).
103 Smooth Endoplasmic Reticulum
Other organelles include the contractile vacuoles of freshwater protozoa that expel excess water that enters cells by osmosis. Some protozoa have extrusomes, vacuoles that release chemicals or structures that deter predators or enable prey capture. A large aqueous central vacuole dominates the volume of many higher plant cells. When filled with water, they will push all other structures against the plasma membrane. In a properly watered plant, this water-filled vacuole exerts osmotic pressure that among other things, keeps plant leaves from wilting and stems upright.
D. Mitochondria and Plastids
Nearly all eukaryotic cells contain mitochondria, shown below.
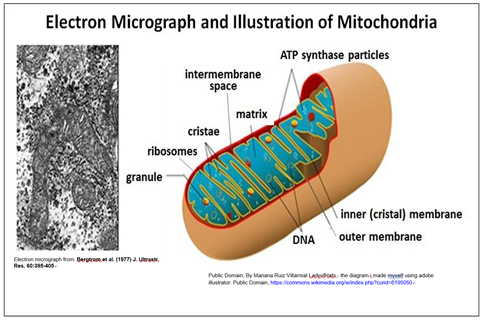
A double membrane surrounds the mitochondrion. Each contains and replicates its own DNA containing genes encoding some mitochondrial proteins. Note that the surface area of the inner mitochondrial membrane is increased by being folded into cristae, which are sites of cellular respiration (aerobic nutrient oxidation).
Earlier, we speculated eukaryotic organelles that could have originated within bacteria. Mitochondria most likely evolved from a complete aerobic bacterium (or proto- bacterium) that was engulfed by a primitive eukaryotic cell. The bacterium escaped destruction, becoming an endosymbiont in the host cell cytoplasm. Lynn Margulis first proposed the Endosymbiotic Theory (Margulis, L. [Sagan, L], 1967. On the origin of mitosing cells. Journal of Theoretical Biology 14 (3): 225–274; available at: Margulis L. Endosymbiotic theory). Margulis proposed that chloroplasts also started as endosymbionts. Like mitochondria, the plastids of plants and some algae have their own DNA, most likely originating as cyanobacteria engulfed by primitive eukaryotic cells. Living in symbiosis with the rest of the cell, they would eventually evolve into plastids, including chloroplasts. Detailed evidence for the Endosymbiotic Theory is discussed elsewhere.
A handful of protozoa were found lacking mitochondria and other organelles. This had suggested they might share ancestry with those primitive eukaryotes that acquired mitochondria by endosymbiosis. However, since such cells contain other organelles such as hydrogenosomes and mitosomes, it is thought more likely that these species once had, but then lost mitochondria. Therefore, descendants of ancient eukaryotic cells missing mitochondria probably no longer exist.
Chloroplasts and cyanobacteria contain chlorophyll and use a similar photosynthetic mechanism to make glucose. A typical chloroplast is shown in the micrograph below (left). A chloroplast beginning to store nutrient sugar as starch is at the right.
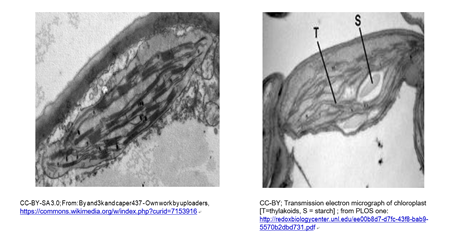
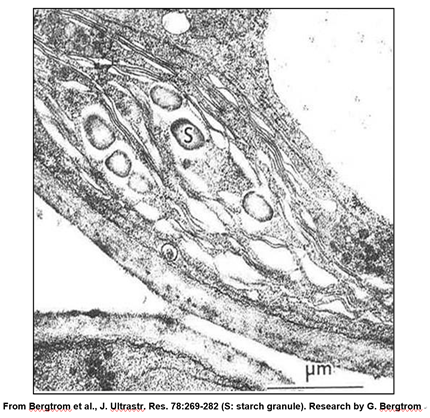
A leucoplast is a plastid a chloroplast that has become filled with starch granules. In the micrograph below, you can see that, because of starch accumulation, the grana have become dispersed and indistinct, forming a leucoplast.
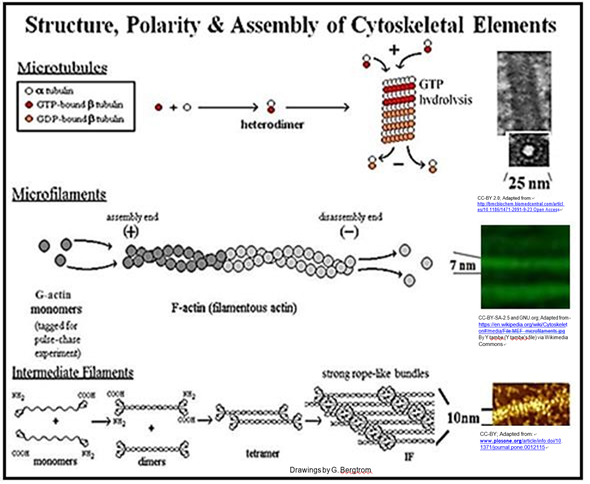
E. Cytoskeletal structures
We have come to understand that the cytoplasm of a eukaryotic cell is highly structured, permeated by rods and tubules. The three main components of this cytoskeleton are microfilaments, intermediate filaments and microtubules.
Microtubules are composed of a- and b-tubulin protein monomers. Monomeric actin proteins make up microfilaments. Intermediate filament proteins are related to keratin, a protein found in hair, fingernails, bird feathers, etc. Cytoskeletal rods and tubules not only determine cell shape, but also play a role in cell motility. This includes the movement of cells from place to place and the movement of structures within cells.
We have already noted that a prokaryotic cytoskeleton is composed in part of proteins homologous to actins and tubulins. As in a eukaryotic cytoskeleton, these bacterial proteins may play a role in maintaining or changing cell shape. On the other hand, flagellum-powered movement in bacteria relies on flagellin, a protein not found in eukaryotic cells. A bacterial flagellum is actually a rigid hook-like structure attached to a molecular motor in the cell membrane that spins to propel the bacterium through a liquid medium.
In contrast, eukaryotic microtubules slide past one another causing a more flexible flagellum to undulate in wave-like motions. Likewise, the motion of a eukaryotic cilium is based on sliding microtubules, in this case causing the cilia to beat rather than undulate. Cilia are involved not only in motility, but also in feeding and sensation. The structures and assembly of the main cytoskeletal components are shown below.
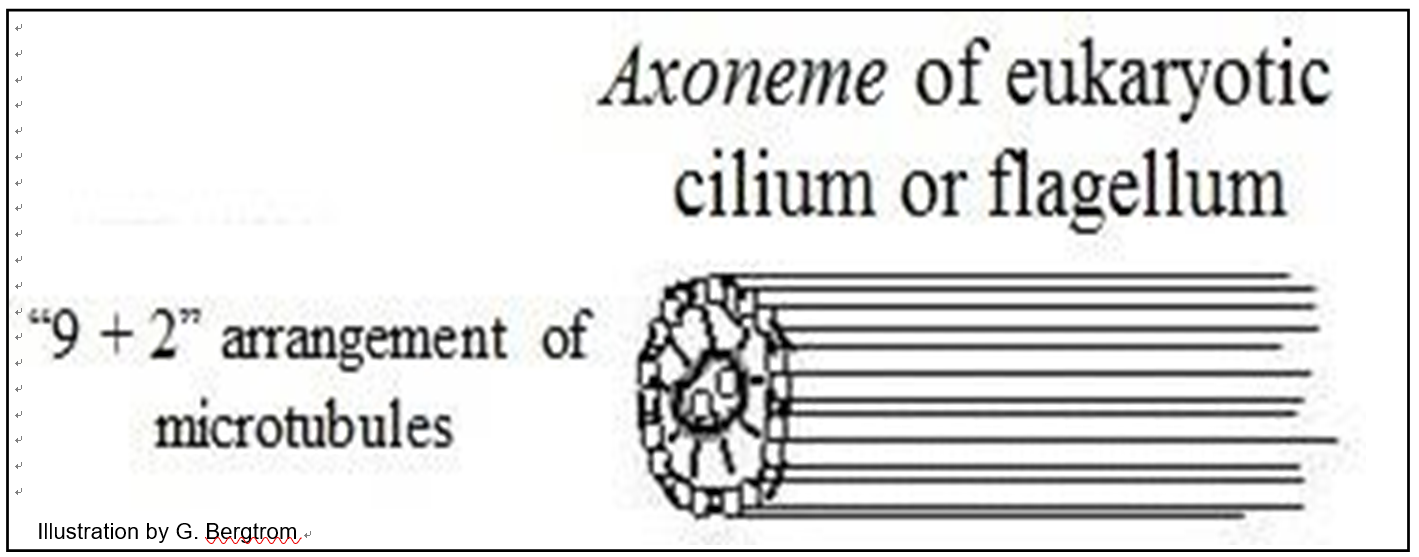
Microtubules in eukaryotic flagella and cilia arise from a basal body (similar to kinetosomes or centrioles). Aligned in a flagellum or cilium, microtubules form an axoneme surrounded by plasma membrane. In electron micrographs of cross sections, a ciliary or flagellar axoneme is typically organized as a ring of nine paired microtubules (called doublets) around two singlet microtubules (illustrated below).
Centrioles are themselves comprised of a ring of microtubules. In animal cells they participate in spindle fiber formation during mitosis and are the point from which microtubules radiate thorough the cell to help form and maintain its shape. These structures do not involve axonemes. The spindle apparatus in plant cells, which typically lack centrioles, form from an amorphous structure called the MTOC, or MicroTubule Organizing Center, which serves the same purpose in mitosis and meiosis as centrioles do in animal cells.
106 Filaments & Tubules of the Cytoskeleton
Elsewhere, we describe how microfilaments and microtubules interact with motor proteins (dynein, kinesin, myosin, etc.) to generate force that results in the sliding of filaments and tubules to allow cellular movement. You will see that motor proteins can also carry cargo molecules from one place to another in a cell.


