26.2: Protein Synthesis
- Page ID
- 15202
Prokaryotic Initiation
The small subunit of the ribosome (the 30S) interprets the genetic information by selecting aminoacyl-tRNAs cognate to the mRNA codons in the decoding center. The large subunit (the 50S) carries the catalytic peptidyl transferase center where amino acids are polymerized into a protein. Small and large subunits unite together at the start codon of a gene to form the 70S ribosome and dissociate again at the stop codon upon completing the synthesis of the encoded protein. This process consists of three phases: initiation, elongation, and termination. In this section, we will focus on the initiation of translation.
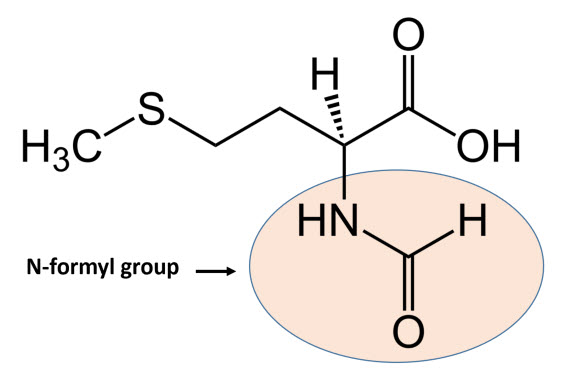
In bacteria, the initiation phase of protein synthesis involves a limited number of “actors”. Aside from the two ribosomal subunits, key roles are played by the initiator tRNAfmet, the translation initiation region (TIR) of the mRNA, and three protein factors – the initiation factors (IFs) IF1, IF2, and IF3 – that ensure speed and accuracy to the overall process. The initiator tRNAfmet contains a methionine residue that has been enzymatically modified to contain an N-terminal formyl group, as shown in Figure \(\PageIndex{1}\). fMet is only used for the initiation of protein synthesis and is thus found only at the N-terminus of the protein. Unmodified methionine is used during the rest translation. Once protein synthesis is completed, the formyl group on methionine may be removed by peptide deformylase, and on occasion, the entire methionine residue can be further removed by the enzyme methionine aminopeptidase.
The TIR sequence within the mRNA contains the start codon and usually an upstream untranslated region that interacts with the small subunit of the ribosome. The bacterial cell produces and expresses a plethora of different mRNAs with different TIR sequences and structures; the efficiency by which these individual transcripts are translated depends not only upon their abundance and stability but also upon the nature of TIR. Thus, unlike the other aforementioned actors that represent constants, the mRNA TIRs represent essentially the only variable in the process of mRNA initiation site selection and can affect translation efficiency.
Although the triplet AUG is by far the most frequent initiation codon found in TIRs, other initiation triplets (i.e., GUG, UUG, AUU, AUC, and AUA) are found in bacteria and the central U is the only universally conserved base of the start codon. Among the aforementioned triplets, those having a 3′-G (i.e., AUG, GUG, and UUG) are recognized equivalently and most efficiently by IF3 during the initiation complex formation.
Another important characteristic of a large number of bacterial mRNA TIRs is the presence of a Shine–Dalgarno (or SD) sequence that is complementary to the 3′ end sequence of 16S rRNA (the anti-SD sequence or aSD). The SD sequence, when present, is usually at an optimal distance of 4–9 nucleotides upstream of the initiation codon, as shown in Figure \(\PageIndex{2}\). While the SD sequence plays an important role in the efficient translation of many mRNA transcripts, it is not essential. Many other mRNA sequences fully lack an SD sequence but are still efficiently transcribed. Thus, the SD sequence is only one example of TIR mechanisms that can play an important role in mRNA binding with the small subunit of the ribosome.

Prokaryotic mRNA sequences often share a highly conserved sequence upstream of the start codon known as the Shine-Dalgarno sequence. This consensus sequence is complimentary to the 3′-end of the 16S rRNA sequence in the small subunit of the ribosome. It is an important feature for the binding and docking of many mRNA molecules with the small ribosomal subunit during transcription initiation.
The three protein initiation factors, IF1, IF2, and IF3, determine the kinetics and fidelity of the overall initiation process. The three IFs are bound, one copy each, to specific sites of the 30S subunit where they assist with the formation of the initiation complex and assembly of the 70S ribosome.
As noted above, the initiator tRNA is first aminoacylated with methionine whose α-NH2 group is eventually blocked by a specific formyl transferase (TMF) to produce a tRNAfmet molecule. This modification prevents interaction with the elongation factor EF-Tu (which we will see plays an important role in the elongation phase of translation, but not the initiation phase!) Blocking EF-Tu binding ensures instead the recognition and binding of tRNAfmet by initiation factor IF2, effectively docking it with the 30S subunit. Furthermore, tRNAfmet binds with high affinity to the ribosomal P-site, unlike all other aminoacyl-tRNAs that bind to the A-site in a ternary complex with EF-Tu and GTP (details will be presented in the next section). In the P-site, the initiator tRNA must be recognized as correct by the other initiation factors IF3 and IF1.
To form the 30S initiation complex, IF3, and IF2 are the first factors to bind to the 30S subunit forming an unstable 30S-IF3-IF2 complex, as shown in Figure \(\PageIndex{3}\) (panel A). The binding of IF1 causes a conformational change in the 30S subunit stabilizing the complex and allowing the recruitment of the tRNAfmet by IF2. Notably, IF1 binds in the A site of the 30S subunit, where it contacts ribosomal protein S12. Recruitment of the tRNAfmet can also stabilize the mRNA interactions with the 30S subunit through the formation of hydrogen bonds between the codon of the mRNA and the anticodon of the tRNAfmet. Note that the binding of mRNA to the 30S subunit is IF-independent and can take place at any time during the 30S assembly process. Two potential routes of mRNA association are shown Figure \(\PageIndex{3}\), panel A, where the mRNA is assembled either prior to or after tRNAfmet recruitment.
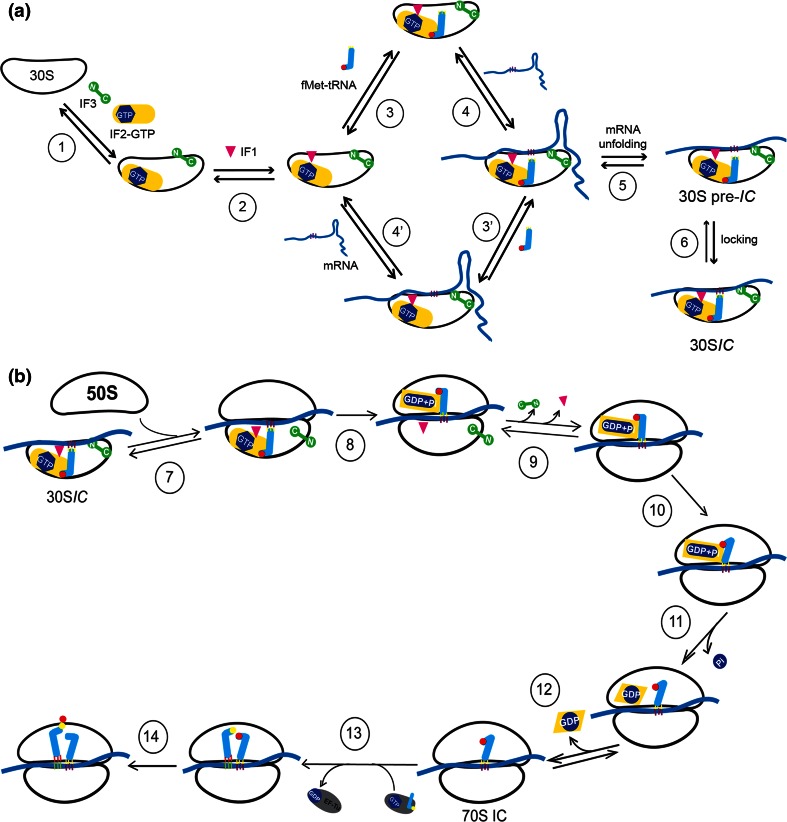
Step 1: a vacant 30S ribosomal subunit binds IF3 and IF2. Step 2: IF1 binds to the 30S subunit in the presence of both IF3 and IF2.
Steps 3 and 3′: in the presence of all three factors tRNAfmet is recruited.
Steps 4 and 4′:the mRNA is bound with different on and off rates depending on its TIR structure; mRNAs with strong secondary structures are bound more slowly than those having little or no secondary structure.
Step 5: mRNAs containing secondary structures must be unfolded in a process that is facilitated by IF2 bound to GTP and antagonized by IF3.
Step 6: the isomerization of the 30S pre-IC allows the P-site codon–anticodon interaction to yield a more stable 30SIC from which mRNA and fMet-tRNA are more stably bound.
Step 7: a 30SIC, containing IF1, IF2·GTP, IF3 and mRNA whose initiation triplet is P-site decoded by fMet-tRNA, is docked by a 50S subunit.
Step 8: upon contact with the 50S subunit, the GTPase function of IF2 is activated and GTP is rapidly hydrolyzed leaving GDP+Pi bound to IF2.
Step 9: this reversible conformational transition represents the last kinetic checkpoint of translation initiation fidelity by IF3 and IF1, as IF3 and IF1 dissociate from the complex.
Step 10: The first-order isomerization of the IF2-GDP structure causes a shift in the ribosome structure that represents the rate-limiting step in 70SIC formation.
Step 11: Pi is dissociated from IF2·GDP.
Step 12: IF2 leaves the ribosome (or moves away from the A-site) clearing the way for EF-Tu binding.
Step 13: the EF-Tu·GTP·aminoacyl-tRNA complex binds to the 70SIC and through a number of steps (not represented here) delivers to the ribosomal A-site the aminoacyl-tRNA encoded by the second mRNA codon.
Step 14:the tRNAfMet bound in the P-site of the peptidyl transferase center donates its formyl-methionine to the A-site-bound aminoacyl-tRNA to yield the initiation dipeptide fMet-aa. Initiation is then complete and the elongation phase can begin.
Following the recruitment of the mRNA and the tRNAfmet to the 30S initiation complex loaded with the IF2, IF3 and IF1 initiation factors, the 50S subunit is very rapidly docked to yield an initially unstable 70S initiation complex (Fig 27.2.3 b). It should be noted that the IF2 protein is a GTP hydrolase enzyme and, as such, binds with the cofactor GTP prior to the recruitment of the 50S subunit. Contact between the IF2 GTPase activating center with the 50S subunit causes the rapid hydrolysis of GTP to GDP + Pi.
The formation of the 70S complex causes the dissociation of the initiation factors. IF2 is the last factor to be dissociated, leaving the ribosome after having positioned tRNAfMet in the P-site of the 70S initiation complex. It must be placed in the correct orientation to facilitate peptide bond formation. GDP and Pi also dissociate from the complex with the removal of IF2. The elongation factor, EF-G is then free to chaperone the first tRNA into the A-site and the first peptide bond is formed (Step 13 of Fig. 27.2.3 b). This marks the beginning of the elongation phase of protein synthesis.
Eukaryotic Initiation
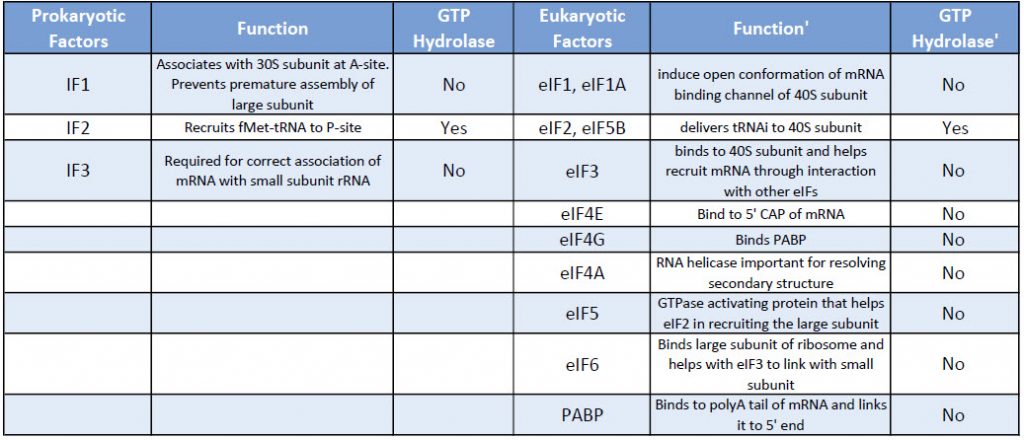
Eukaryotic translation initiation is more complex than prokaryotic systems and requires the actions of at least 11 eukaryotic initiation factors (eIFs), plus additional auxiliary factors (Table \(\PageIndex{1}\)). We will not cover the action of all these eIFs in detail here, but rather focus a few key steps as outlined in Figure \(\PageIndex{4}\).
Table \(\PageIndex{1}\): Comparison of Prokaryotic and Eukaryotic Translation Initiation Factors
First, the initiator tRNAi is recruited to the small ribosomal subunit (40S) to form a ternary complex with the GTP-bound eukaryotic initiation factor 2 (eIF2). Formation of this 43S pre-initiation complex is strongly enhanced by additional factors, such as eIF3. eIF3 also interacts with the eIF4F complex, which consists of three other initiation factors: eIF4A, eIF4E, and eIF4G. eIF4G is a scaffolding protein that directly associates with both eIF3 and the other two components. eIF4E is the 5′-cap-binding protein. Binding of the mRNA cap by eIF4E is often considered the rate-limiting step of cap-dependent initiation, and the concentration of eIF4E is a regulatory nexus of translational control. Certain viruses cleave a portion of eIF4G that binds eIF4E, thus preventing cap-dependent translation to hijack the host machinery in favor of the viral (cap-independent) messages. eIF4A is an ATP-dependent RNA helicase that aids the ribosome by resolving certain secondary structures formed along the mRNA transcript. The poly(A)-binding protein (PABP) also associates with the eIF4F complex via eIF4G, and binds the poly-A tail of most eukaryotic mRNA molecules. This protein has been implicated in playing a role in circularization of the mRNA during translation.The 43S preinitiation complex accompanied by the protein factors moves along the mRNA chain toward its 3′-end, in a process known as ‘scanning’, to reach the start codon (typically AUG). After recognition of the start codon, the large ribosomal subunit (60S) assembles to form the 80S initiation complex, which is ready for elongation. Alternatively, under distinct conditions or on certain transcripts internal initiation can occur in a cap-independent manner at so called internal ribosome entry sites (IRES). Eukaroytic translation initiation is shown in Figure \(\PageIndex{4}\).
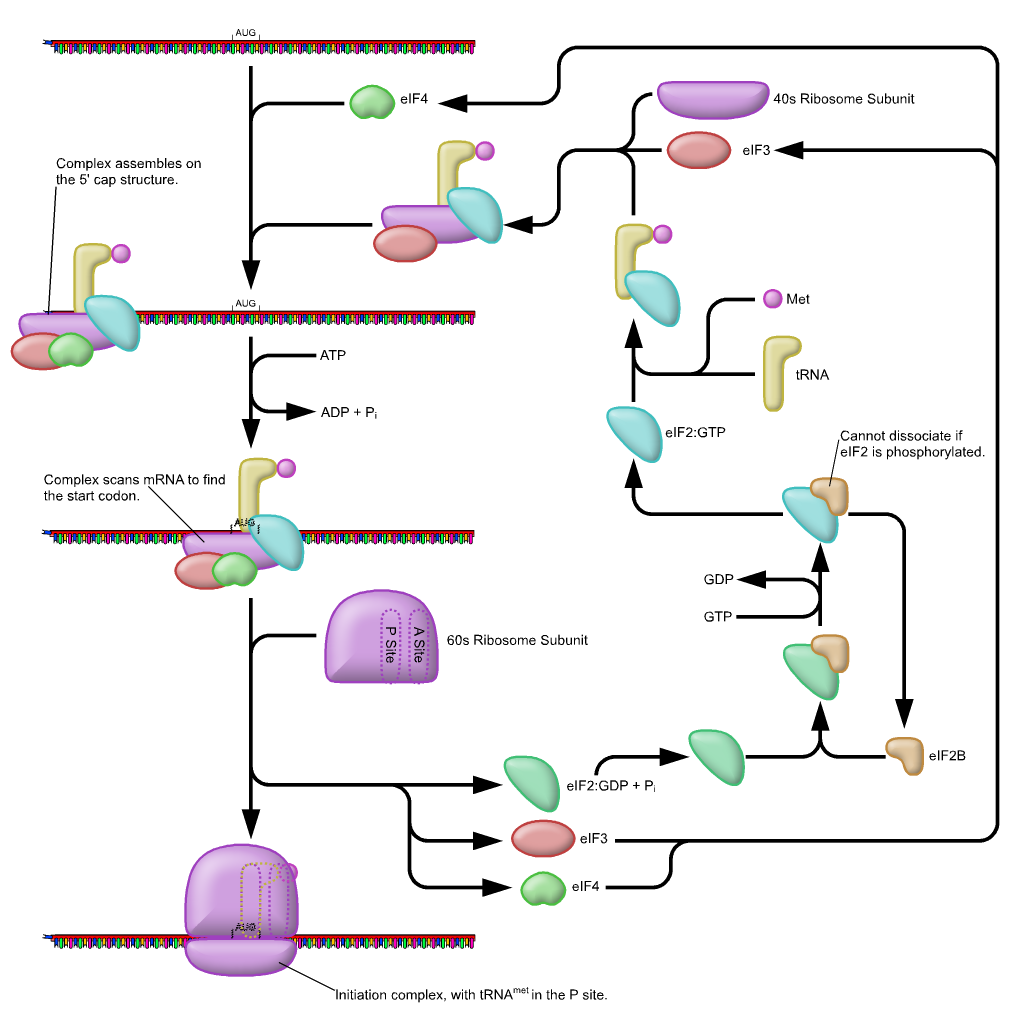
This is a simplified diagram of eukaryotic translation initiation detailing some of the eIFs involved in the process. eIF2 is critical for recruiting the initiation tRNAi to the 40S subunit. eIF3 enhances the activity of eIF2 and also promotes the binding of the 43S pre-initiation complex with the mRNA. eIF3 binds with the mRNA through the interaction of the eIF4 factors and causes the scanning of the pre-initiation complex down the mRNA to locate the start codon (usually AUG). Poly A Binding Proteins (PABPs) bind with the polyA tail sequence of the mRNA and also interact with the eIF4 factors causing the circularization of the mRNA.
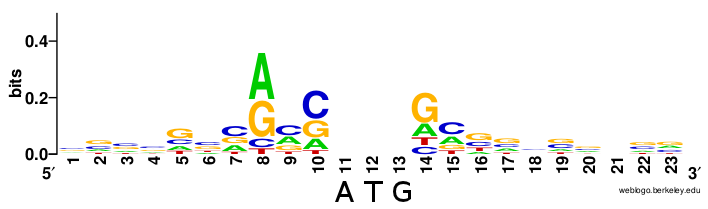
As seen in prokaryotic systems with the favored Shine Dalgarno sequence upstream of the start codon within the mRNA sequence, there are also preferred nucleotide sequences within the local vicinity of the start codon in eukaryotic mRNAs, as well. In eukaryotic mRNA, this is known as the Kozak sequence (Figure \(\PageIndex{5}\)). The sequence was originally defined as 5′-(gcc)gccRccAUGG-3 where:
- The underlined nucleotides indicate the translation start codon, coding for Methionine.
- upper-case letters indicate highly conserved bases, i.e. the ‘AUGG’ sequence is constant or rarely, if ever, changes.
- ‘R’ indicates that a purine (adenine or guanine) is always observed at this position (with adenine being more frequent according to Kozak rules)
- a lower-case letter denotes the most common base at a position where the base can nevertheless vary
- the sequence in parentheses (gcc) is of uncertain significance.
The AUG is the initiation codon encoding a methionine amino acid at the N-terminus of the protein. (Rarely, GUG is used as an initiation codon, but methionine is still the first amino acid as it is the met-tRNA in the initiation complex that binds to the mRNA). Variation within the Kozak sequence alters the “strength” of the translational start site. Kozak sequence strength refers to the favor ability of initiation, affecting how much protein is synthesized from a given mRNA. This is shown in Figure \(\PageIndex{5}\).
The Elongation Phase of Translation
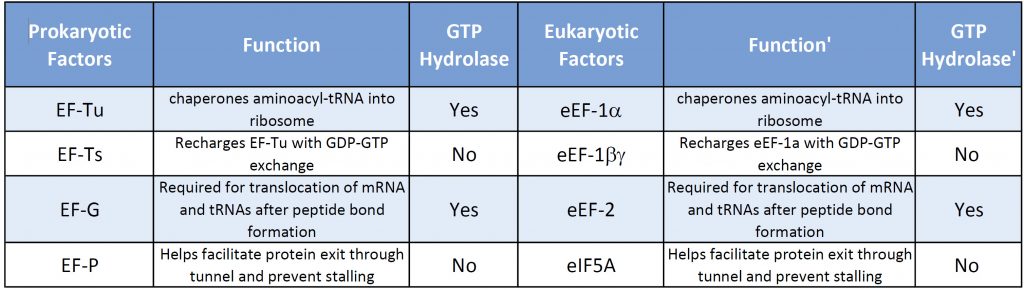
Both prokaryotic and eukaryotic elongation phases of transcription utilize similar elongation factors during the process. Table \(\PageIndex{2}\) provides a summary of their functions.
Table \(\PageIndex{2}\): Comparison of Prokaryotic and Eukaryotic Translation Elongation Factors
Prokaryotic Elongation
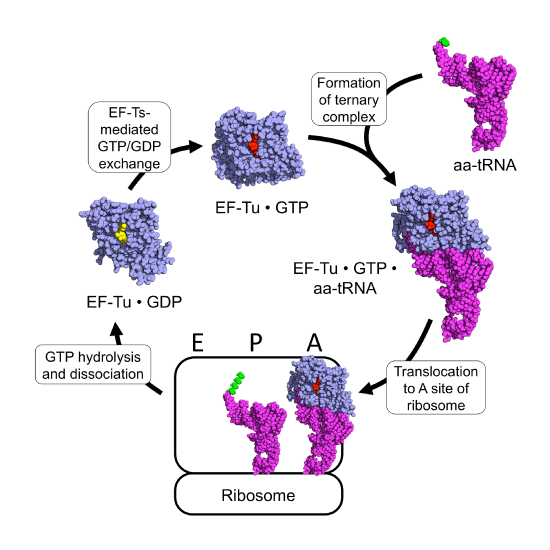
The prokaryotic elongation phase of transcription requires the activity of three primary elongation factors (EFs), EF-Tu, EF-Ts, and EF-G. During elongation, aminoacyl-tRNAs are delivered to the ribosome in the form of a ternary complex: the tRNA, a translational GTPase (in bacteria: EF-Tu or SelB), and a GTP molecule, as shown in Figure \(\PageIndex{6}\). The tRNA decodes the information on the mRNA by forming hydrogen bonds (H-bonds) between codon and anticodon nucleobases. Remarkably, the free-energy difference between correct (cognate) and incorrect (near-cognate, non-cognate) base pairing alone does not explain the very high fidelity of decoding. Rather, high fidelity is achieved by a two-step decoding process: initial selection leading to GTPase activation and proofreading. In addition to the free-energy difference, kinetic effects contribute to the discrimination. The GTP hydrolysis rate is increased and tRNA rejection rate is decreased by the recognition of the correct codon.
Small-subunit nucleotides A1492 and A1493 adopt a flipped-out conformation in the presence of a tRNA and, in this conformation, the tRNA anticodon hydrogen bonds with the codon of the mRNA forming a mini-helix structure as shown in Figure \(\PageIndex{7}\). The flipped out nucleotides A1492, A1493 along with G530 were found to shield the codon–anticodon base pairs from solvent. This shielding of near-cognate base pairs from the solvent is incomplete causing an increase in the free-energy difference between near-cognate and cognate base pairs and more flexibility within the docking region. This reduces the strength of hydrogen bonding between a non-cognate tRNA and causes the inappropriate tRNA to leave the A-site before peptide bond formation can occur. This increases the fidelity and discrimination of tRNA selection, such that only the correct cognate tRNA is incorporated into the A-site.
Interestingly aminoglycosides, a class of antibiotics, bind to the decoding center and lock nucleotides A1492/A1493 in the flipped-out conformation as shown in Figure \(\PageIndex{8}\). In this way aminoglycosides promote the accommodation of near-cognate, thus wrong, tRNAs into proteins during synthesis causing wide-spread mutagenesis. This is toxic to the bacteria and leads to bacterial cell death.
Figure \(\PageIndex{8}\): Structure of the Aminoglycoside Antibiotic, Puromycin. Figure from: Yikrazuul
After GTP hydrolysis, the GTPase EF-Tu dissociates from the tRNA. At this point, the EF-Tu is tightly bound with a molecule of GDP and cannot release GDP on its own to be recycled for a second round of tRNA chaperoning. The recharging of EF-Tu is executed by the Elongation Factor Thermo stable (EF-Ts), as shown in Figure \(\PageIndex{9}\). The binding of EF-Ts with EF-Tu-GDP causes a conformational change in EF-Tu that allows the release of GDP. The binding of a new molecule of GTP with the EF-Tu protein causes the dissociation of EF-Ts and fully recharges EF-Tu.
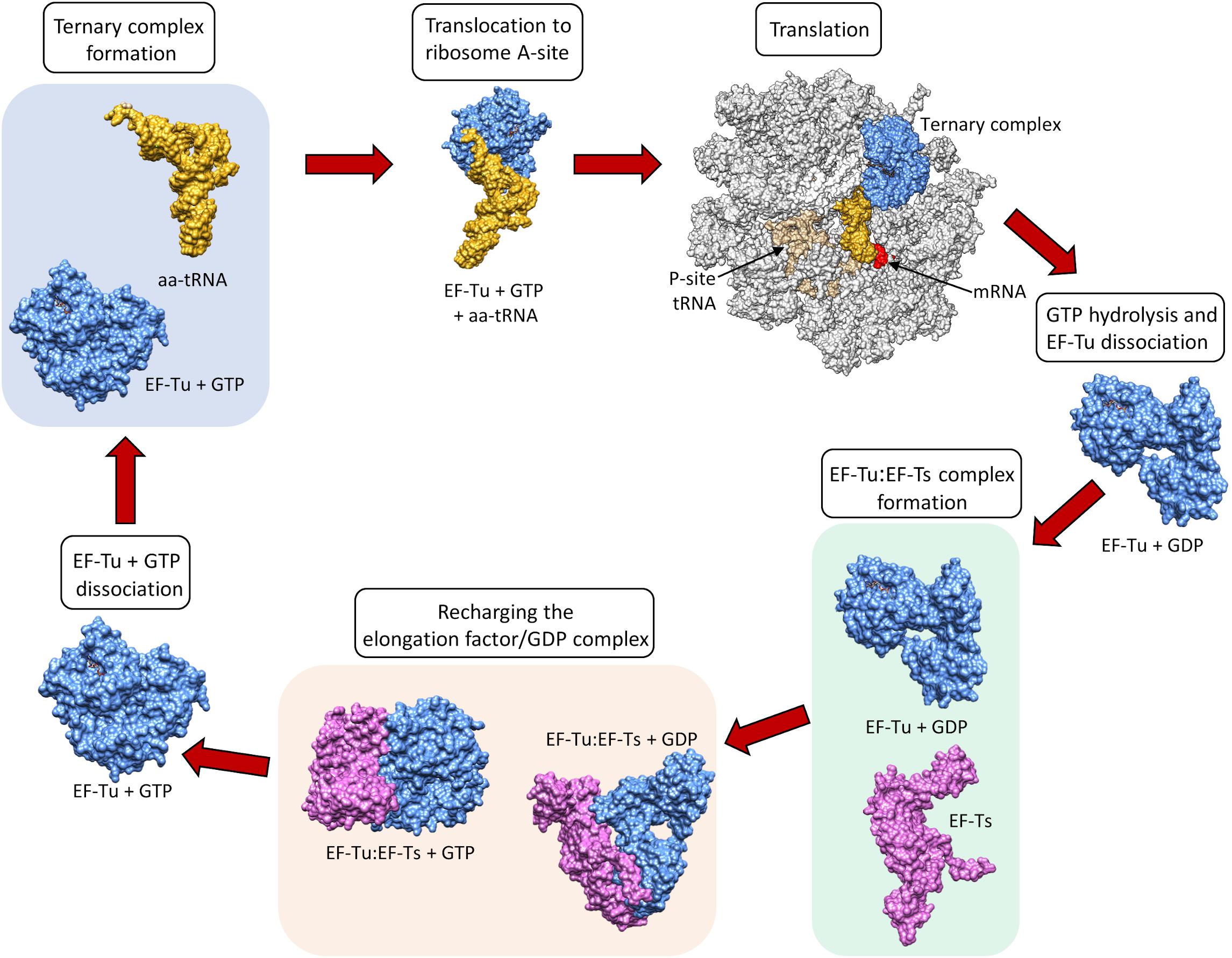
Dissociation of EF-Tu from the ribosome allows the tRNA to move into to the peptidyl transferase center (A-site) on the large subunit. At the core of ribosomal translation is the catalysis of peptide bond formation, as shown in Figure \(\PageIndex{10}\). The current reaction models point to a substrate assisted mechanism. Simulations indicate that the transition state forms due to extensive hydrogen bonding with water molecules and the surrounding rRNA bases and that the C-O bond cleavage takes place after C-N bond formation. Peptide bond formation results in the transition of the amino acid docked on the P-site tRNA to the nascent growing peptide that is now held on the tRNA in the A-site. Note that this mechanism causes the nascent growing peptide to always grow in the N- to C- direction.
Once the peptide bond is formed, the ribosome needs to translocate down the mRNA to make the next mRNA codon available within the A-site. This also requires the shifting of the tRNA molecules, such that the tRNA in the A-site (which is now tethered to the nascent peptide) shifts to the P-site. The P-site tRNA (which is now empty) shifts to the E-site, and if there was an empty tRNA in the E-site, it will shift to exit the ribosome. Shifting the tRNAs and mRNAs within the ribosome core requires the action of the EF-G elongation factor, as shown in Figure \(\PageIndex{11}\).
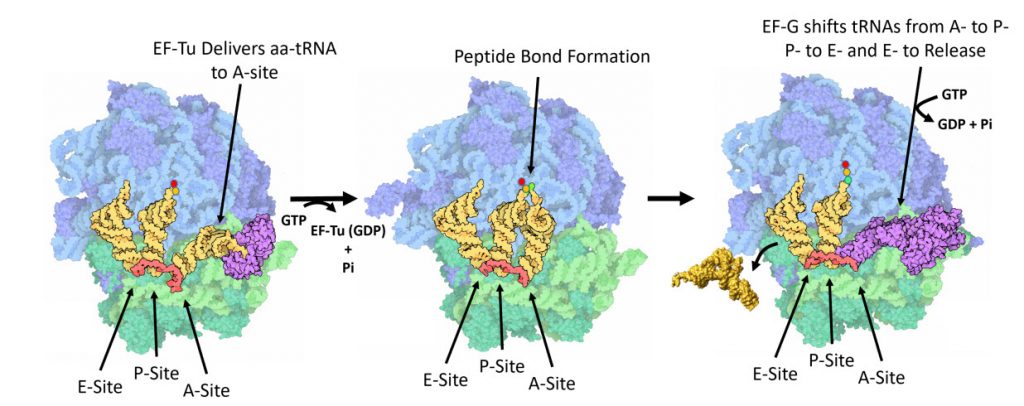
Figure \(\PageIndex{12}\) shows an interactive iCn3D model of the eukaryotic 80S ribosome with bound mRNA and tRNAs (6GX3) . (Very long load time)
.png?revision=1&size=bestfit&width=489&height=499)
Figure \(\PageIndex{12}\): eukaryotic 80S ribosome with bound mRNA and tRNAs (6GX3) . (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...Ak7ZvtRxaoAQp9 (Very long load time)
color coding as follows:
- gray: protein tube
- coiled coils: RNA trace
- black spheres: mRNA
- dark blue spheres: ap/P-site tRNA
- cyan spheres: pe/E-site-tRNA
- green spheres: Mg2+
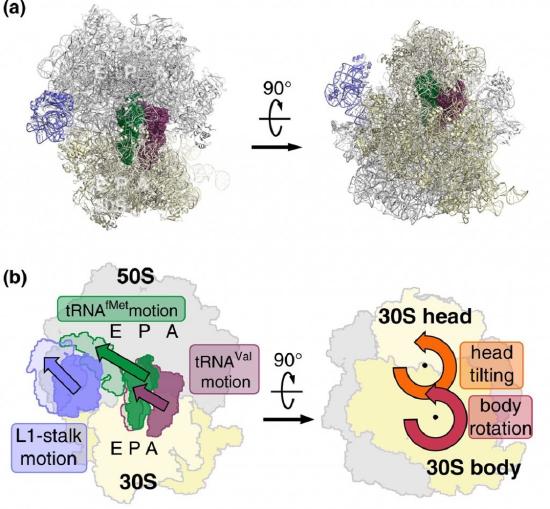
EF-G is a GTP hydrolase protein that binds to the A-site of the ribosome. The EF-G protein has high flexibility that enables it to act as a hinge. Folding of EF-G is dependent on GTP hydrolysis. Thus, when binding to the ribosome, the fast hydrolysis of GTP acts as a power stroke folding the EF-G protein and causing a conformation shift in the ribosome that enables the translocation of the tRNA residues and the mRNA. Translocation of tRNAs is accompanied by large-scale collective motions of the ribosome: relative rotation of ribosomal subunits and L1-stalk motion, as shown in Figure \(\PageIndex{13}\). The L1 stalk, which is a flexible part of the large subunit, is in contact and moves along with the tRNA from the P to the E site. Once in the EF-G-GDP form, the factor quickly dissociates from the ribosome, opening up the A-site for the recruitment of the next aa-tRNA molecule. The elongation cycle will continue to be repeated until a termination codon is reached.
Eukaryotic Elongation
The elongation phase in eukaryotic translation is very similar to prokaryotic elongation. Essentially, the mRNA is decoded by the ribosome in a process that requires the selection of each aminoacyl-transfer RNA (aa-tRNA), which is dictated by the mRNA codon in the ribosome acceptor (A) site, peptide bond formation and movement of both tRNAs and the mRNA through the ribosome, as shown in Figure \(\PageIndex{14}\). A new amino acid is incorporated into a nascent peptide at a rate of approximately one every sixth of a second. The first step of this process requires guanosine triphosphate (GTP)-bound eukaryotic elongation factor 1A (eEF1α) to recruit an aa-tRNA to the aminoacyl (A) site, which has an anticodon loop cognate to the codon sequence of the mRNA. The anticodon of this sampling tRNA does not initially base-pair with the A-site codon. Instead, the tRNA dynamically remodels to generate a codon-anticodon helix, which stabilizes the binding of the tRNA-eEF1α-GTP complex to the ribosome A site. This helical structure is energetically favorable for cognate or correct pairing, and so discriminates between the non-cognate or unpaired and single mismatched or near-cognate species. This is important for the accuracy of decoding since it provides a mechanism to reject a non-cognate tRNA that carries an inappropriate amino acid. The pairing of the tRNA and codon induces GTP hydrolysis by eEF1α, which is then evicted from the A site. In parallel with this process, the ribosome undergoes a conformational change that stimulates contact between the 3′ end of the aa-tRNA in the A site and the tRNA carrying the polypeptide chain in the peptidyl (P) site. The shift in position of the two tRNAs [A to the P site and P to the exit (E) site] results in ribosome-catalyzed peptide bond formation and the transfer of the polypeptide to the aa-tRNA, thus extending the polypeptide by one amino acid. The second stage of the elongation cycle requires a GTPase, eukaryotic elongation factor 2 (eEF2), which enters the A-site and, through the hydrolysis of GTP, induces a change in the ribosome conformation. This stimulates ribosome translocation to allow the next aa-tRNA to enter the A-site, thus starting a new cycle of elongation.
Figure \(\PageIndex{14}\): Eukartyotic Translation Elongation Phase. Knight, J.R.P., et. al. (2020) Disease Models & Mechanisms 13, dmm043208.
This schematic represents the four basic steps of eukaryotic translation elongation. The ribosome contains three tRNA-binding sites: the aminoacyl (A), peptidyl (P) and exit (E) sites. In the first step of peptide elongation, the tRNA, which is in a complex with eIF1 and GTP and contains the cognate anticodon to the mRNA coding sequence, enters the A site. Recognition of the tRNA leads to the hydrolysis of GTP and eviction of eEF1 from the A site. In parallel, the deacylated tRNA in the E site is ejected. The A site and the P site tRNAs interact, which allows ribosome-catalyzed peptide bond formation to take place. This involves the transfer of the polypeptide to the aa-tRNA, thus extending the nascent polypeptide by one amino acid. eIF5A allosterically assists in the formation of certain peptide bonds, e.g. proline-proline. eEF2 then enters the A site and, through the hydrolysis of GTP, induces a change in the ribosome conformation and stimulates translocation. The ribosome is then in a correct conformation to accept the next aa-tRNA and commence another cycle of elongation.
The Ribosome as a Ribozyme
Protein synthesis from a mRNA template occurs on a ribosome, a nanomachine composed of proteins and ribosomal RNAs (rRNA). Peptide bond formation occurs when another tRNA-amino acid molecule binds to an adjacent codon on mRNA. The tRNA has a cloverleaf tertiary structure with some intrastranded H-bonded secondary structure. The last three nucleotides at the 3' end of the tRNA are CpCpA. The amino acid is esterified to the terminal 3'OH of the terminal A by a protein enzyme, aminoacyl-tRNA synthetase.
Covalent amide bond formation between the second amino acid to the first, forming a dipeptide, occurs at the peptidyl transferase center, located on the larger ribosomal subunit (50S and 60S in bacteria and eukaryotes, respectively). The ribosome ratchets down the mRNA so the dipeptide-tRNA is now at the the P or Peptide site, awaiting a new tRNA-amino acid at the A or Amino site. Figure \(\PageIndex{15}\): below shows a schematic of the ribosome with bound mRNA on the 30S subunit and tRNAs covalently attached to amino acid (or the growing peptide) at the A and P site, respectively.
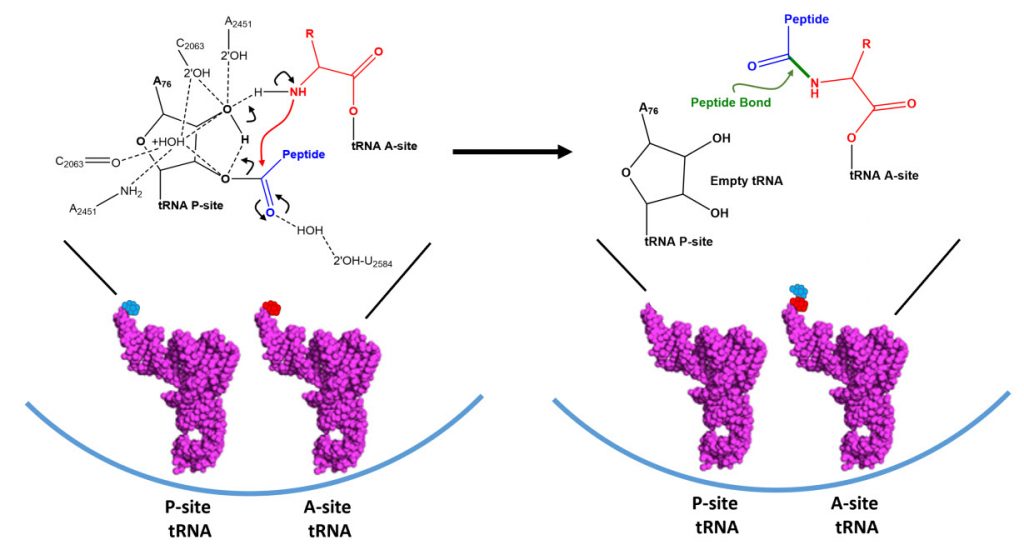
A likely mechanism (derived from crystal structures with bound substrates and transition state analogs) for the formation of the amide bond between a growing peptide on the P-site tRNA and the amino acid on the A-site tRNA is shown in Figure \(\PageIndex{16}\). Catalysis does not involve any of the ribosomal proteins (not shown) since none is close enough to the peptidyl transferase center to provide amino acids that could participate in general acid/base catalysis, for example. Hence the rRNA must act as the enzyme (i.e. it is a ribozyme). Initially it was thought that a proximal adenosine with a perturbed pKa could, at physiological pH, be protonated/deprotonated and hence act as a general acid/base in the reaction. However, none was found. The most likely mechanism to stabilize the oxyanion transition state at the electrophilic carbon attack site is precisely located water, which is positioned at the oxyanion hole by H-bonds to uracil 2584 on the rRNA. The cleavage mechanism involves the concerted proton shuffle shown below. In this mechanism, the substrate (Peptide-tRNA) assists its own cleavage in that the 2'OH is in position to initiate the protein shuttle mechanism. (A similar mechanism might occur to facilitate hydrolysis of the fully elongated protein from the P-site tRNA.) Of course all of this requires perfect positioning of the substrates and isn't that what enzymes do best? The main mechanisms for the catalysis of peptide bond formation by the ribosome (as a ribozyme) are intramolecular catalysis and transition state stabilization by the appropriately positioned water molecule.
Translation Termination
Prokaryotic Termination
Termination of bacterial protein synthesis occurs when a stop codon is presented in the ribosomal A-site and is recognized by a class I release factor, RF1 or RF2. These release factors (RFs) have different but overlapping specificities, where RF1 reads UAA and UAG and RF2 reads UAA and UGA, with strong discrimination against sense codons. The RFs are multi-domain proteins, where binding and stop codon recognition by domain 2 at the decoding site causes the universally conserved GGQ motif of domain 3 to insert into the A-site of the PTC, some 80 Å away from the decoding site. This event triggers hydrolysis of the peptidyl-tRNA bond in the P-site of the PTC, and the nascent peptide chain can then be released via the ribosomal exit tunnel, as shown in Figure \(\PageIndex{14}\). After peptide release, RF1 and RF2 dissociate from the post-termination complex. The dissociation is accelerated by a class II release factor called RF3, which functions as a translational GTPase that binds and hydrolyses GTP in the course of termination.
While RF3 increases the efficiency of peptide hydrolysis, it is not an essential protein for the process. In gene knockout studies, RF3 is dispensable for growth of Escherichia coli, and its expression is not conserved in all bacterial lineages. For example, RF3 is not present in the thermophilic model organisms of the Thermus and Thermatoga genera and in infectious Chlamydiales and Spirochaetae. This means that both RF1 and RF2 are capable of performing a complete round of termination independently of RF3 or that other GTPases from the elongation or initiation phases of translation can compensate for the action of RF3.
The release factors RF1 and RF2 acquire an open conformation (Figure \(\PageIndex{17}\) on the 70S ribosome, which is distinctly different from the closed conformation observed in crystal structures of free RFs. The conformational equilibrium of the free RFs in solution shows that this open conformation is dominating at about 80%.
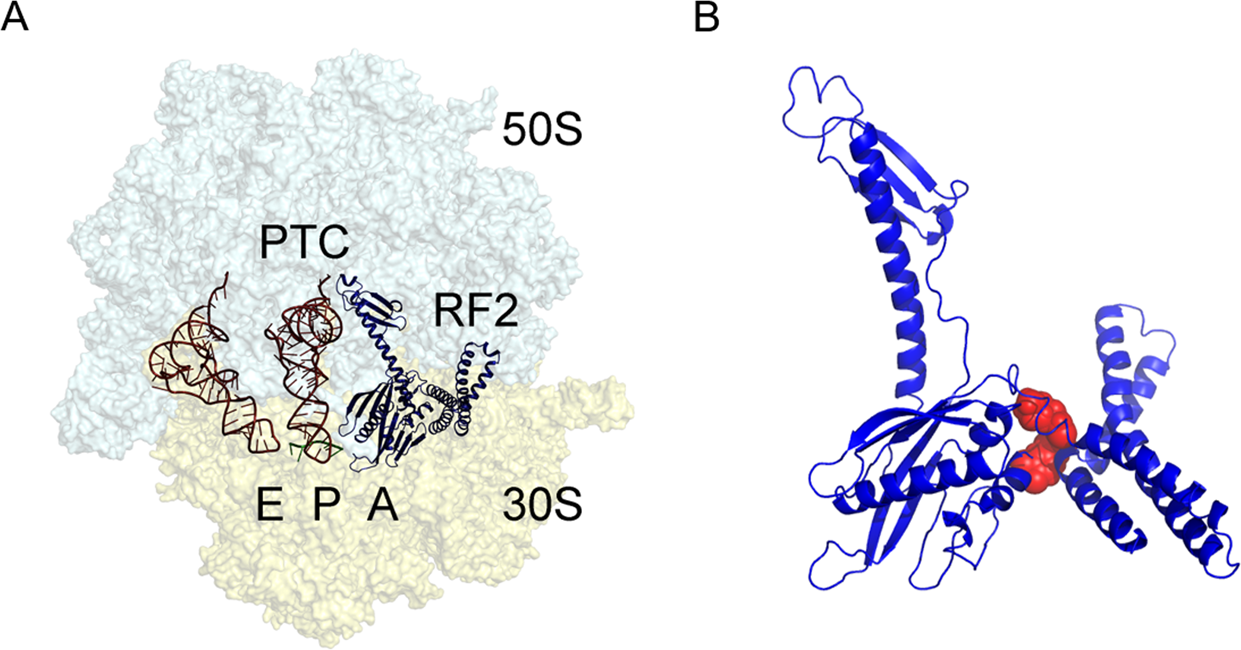
During peptide hydrolysis, the RF factors cause rotational and conformational changes within the ribosome that allow the binding of a ribosome recycling factor (RRF) and the EF-G GTPase, which leads to the dissociation of the large subunit from the small subunit and the release of the mRNA, as shown in Figure \(\PageIndex{18}\).
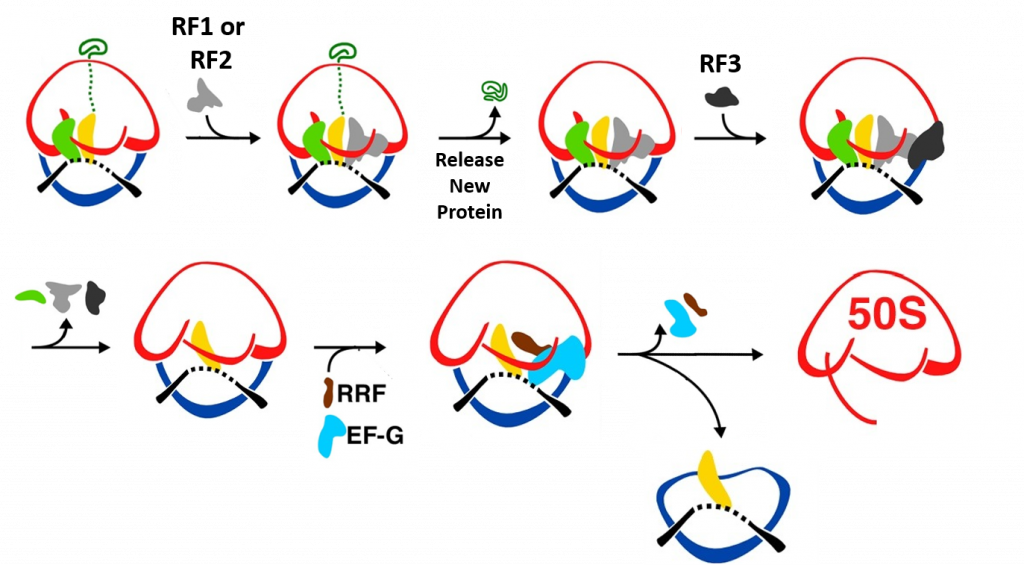
When a stop codon enters the A-site of the ribosome RF1 or RF2 enter the A-site and bind with the mRNA. This leads to the hydrolysis of the protein and release through the exit tunnel. Binding of RF3 and GTP hydrolysis causes the dissociation of the RF factors and conformational change of the ribosome structure. Subse
Eukaryotic Termination
In eukaryotes and archaea, on the other hand, a single omnipotent RF reads all three stop codons. Although the mechanism of translation termination is basically the same, there is neither sequence nor structural homology between the bacterial RFs and the eukaryotic eRF1, apart from the universally conserved GGQ motif which is required for peptide hydrolysis from the tRNA. the eRF3 GTPase coordinates the release of eRF1 following hydrolysis. In Archaea, there is no eRF3 homolog, instead the aEF1A protein mediates this function. The process of eukaryotic ribosomal disassembly and recycling is currently not well understood, but appears to involve an ABC type ATPase called ABCE1. Mitochondria have independent RFs that can recognize standard and non-standard stop codons, and are more homologous with bacterial systems of ribosomal recycling and disassembly.
Summary of Translation
An overall summary of prokaryotic translation is given in Figure \(\PageIndex{19}\).
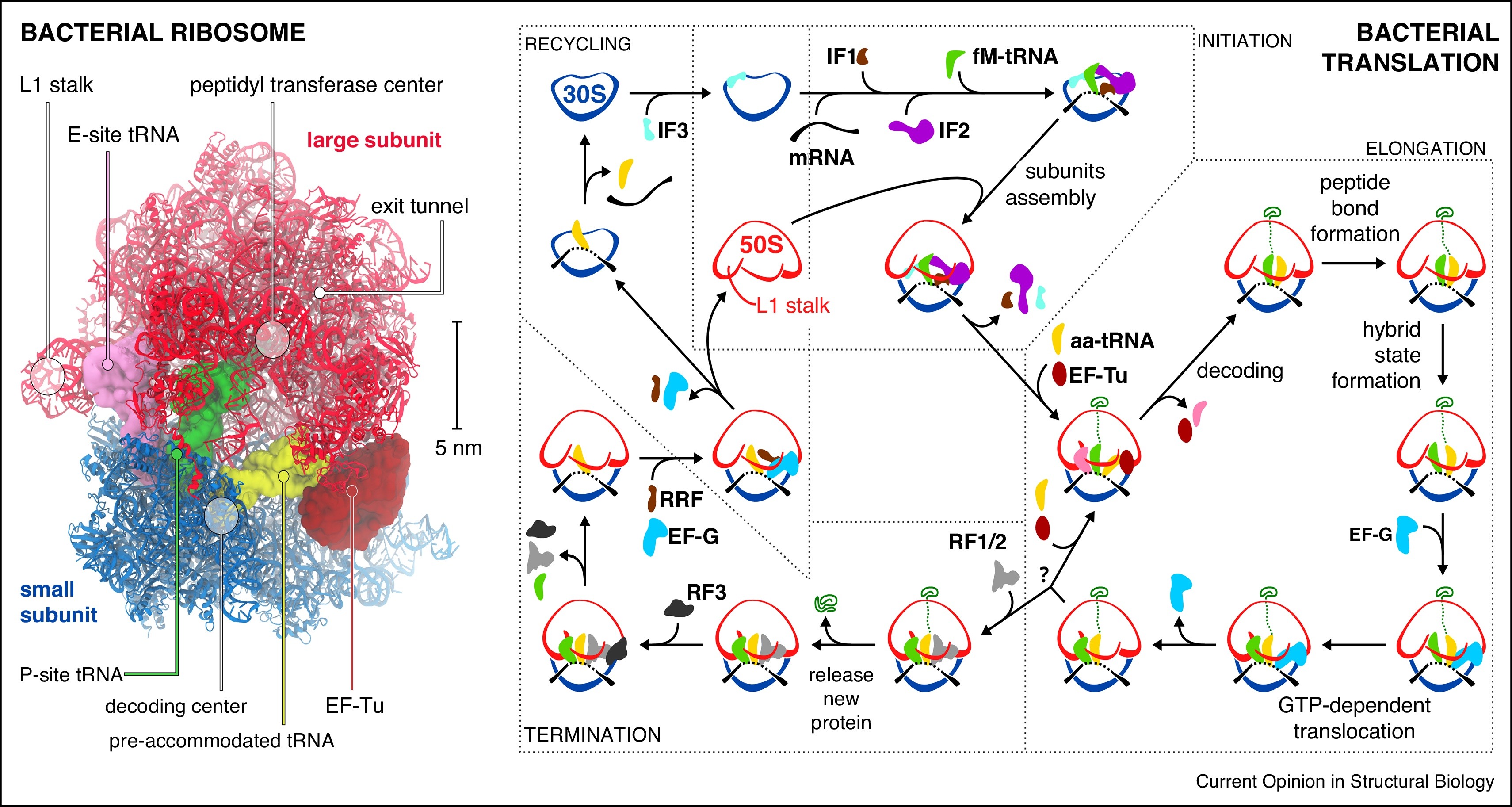
Left panel: Structure of the bacterial ribosome in complex with EF-Tu (PDB 5AFI).
Right Panel: Scheme of the bacterial translation cycle. 30S: small subunit; 50S: large subunit; IF1, IF2, IF3: initiation factors; fM-tRNA: N-formylmethionine tRNA; aa-tRNA: aminoacyl tRNA; EF-Tu, EF-G: elongation factors; RF1, RF2, RF3: release factors; RRF: ribosome recycling factor; green trace: nascent protein. The question mark stands for a stop codon recognition.
References
This chapter was remixed and adapted from the following resources under creative commons licensing:
- Wikipedia contributors. (2020, June 27). Wobble base pair. In Wikipedia, The Free Encyclopedia. Retrieved 16:47, August 12, 2020, from https://en.Wikipedia.org/w/index.php?title=Wobble_base_pair&oldid=964760055
- Lorenz, C., Lünse, C., and Mörl, M. (2017) tRNA modifications: Impact on structure and thermal adaptation. Biomolecules 7(2)35. Available at: https://www.mdpi.com/2218-273X/7/2/35/htm
- Pan, T. (2018) Modifications and functional genomics of human transfer RNA. Cell Research 28:395-404. Available at: https://www.nature.com/articles/s41422-018-0013-y#Sec3
- Bednárová, A., Hanna, M., Durham, I., Van Cleave, T., England, A., Chaudhuri, A., and Krishnan, N. (2017) Lost in translation: Defects in transfer RNA modifications and neurological disorders. Front. Mol Neurosci. 10:135. Available at: https://www.researchgate.net/publication/316440980_Lost_in_Translation_Defects_in_Transfer_RNA_Modifications_and_Neurological_Disorders
- Wikipedia contributors. (2020, May 17). Transfer RNA. In Wikipedia, The Free Encyclopedia. Retrieved 23:03, August 15, 2020, from https://en.Wikipedia.org/w/index.php?title=Transfer_RNA&oldid=957227343
- Gomez, M.A.R., and Ibba M. (2020) Aminoacyl-tRNA Synthetases. RNA, doi: 10.1261/rna.071720.119 Available at: https://rnajournal.cshlp.org/content/early/2020/04/17/rna.071720.119.abstract
- Li, R., Macnamara, L.M., Leuchter, J.D., Alexander, R.W., and Cho, S.S. (2015) MD Simulations of tRNA and Aminoacyl-tRNA Syntetases: Dynamics, Folding, Binding, and Allostery. Int. J. Mol Sci. 16(7):15872-15902. Available at: https://www.mdpi.com/1422-0067/16/7/15872
- Wikipedia contributors. (2020, August 15). Ribosome. In Wikipedia, The Free Encyclopedia. Retrieved 05:22, August 16, 2020, from https://en.Wikipedia.org/w/index.php?title=Ribosome&oldid=973144885
- Kater, L., Thoms, M., Barrio-Garcia, C., Cheng, J., Ismail, S., Ahmed, Y.L., Bange, G., Kressler, D., Berninghausen, O., Sinning, I., Hurt, E., and Beckmann, R. (2017) Visualizing the assembly pathway of nucleolar Pre-60S Ribosomes. Cell 171(7):1599-1610. Available at: https://www.sciencedirect.com/science/article/pii/S0092867417314290
- Bock, L.V., Kolár, M.H., Grubmüller, H. (2018) Molecular simulations of the ribosome and associated translation factors. Cur. Op. Struc. Bio. 49:27-35. Available at: https://www.sciencedirect.com/science/article/pii/S0959440X1730132X
- Doris, S.M., Smith, D.R., Beamesderfer, J.N., Raphael, B.J., Nathanson, J.A., and Gerbi, S.A. (2015) Universal and domain-specific sequences in 23S-28S ribosomal RNA identified by computational phylogenetics. RNA 21:1719-1730. Available at: https://www.researchgate.net/publication/281141702_Universal_and_domain-specific_sequences_in_23S-28S_ribosomal_RNA_identified_by_computational_phylogenetics
- Aleksashin, M.A., Leppik, M., Hochenberry, A.J., Klepacki, D., Vázquez-Laslop, N., Jewett, M.C., Remme, J., and Mankin A.S. (2019) Assembly and functionality of the ribosome with tethered subunits. Nature Communications 10:930. Available at: https://www.nature.com/articles/s41467-019-08892-w#rightslink
- Wikipedia contributors. (2020, July 3). Formylation. In Wikipedia, The Free Encyclopedia. Retrieved 22:03, August 16, 2020, from https://en.Wikipedia.org/w/index.php?title=Formylation&oldid=965827476
- Gualerzi, C.O., and Pon C.L. (2015) Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell Mol Life Sci. 72:4341-4367. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4611024/
- Wikipedia contributors. (2020, June 14). Kozak consensus sequence. In Wikipedia, The Free Encyclopedia. Retrieved 06:03, August 18, 2020, from https://en.Wikipedia.org/w/index.php?title=Kozak_consensus_sequence&oldid=962444929
- Knight, J.R.P., Garland, G., Pöyry, T., Mead, E. Vlahov, N., Sfakianos, A., Frosso, S., De-Lima-Hedayioglu, F., Mallucci, G.R., von der Haar, T., Smales, C.M., Sansom, O.J., and Willis, A.E. (2020) Control of translation elongation in health and disease. Dis. Mod. and Mech. 13: dmm043208. Available at: https://dmm.biologists.org/content/13/3/dmm043208
- Adio, S., Sharma, H., Senyushkina, T., Karki, P., Maracci, C., Wohlgemuth, I., Holtkamp, W., Peske, R., and Rodina, M.V. (2018) Dynamics of ribosomes and release factors during translation termination in E. coli. eLife 7:e34253. Available at: https://elifesciences.org/articles/34252
- Ge, X., Oliveira, A., Hjort, K., Bergfors, T., Guitiérrez-de-Terán, H., Andersson, D.I., Sanyal, S., and Åqvist, J. (2019) Inhibition of translation termination by small molecules targeting ribosomal release factors. Scientific Reports 9: 15424. Available at: https://www.nature.com/articles/s41598-019-51977-1#rightslink
- Svidritskiy, E., Demo G., Loveland A.B., Xu, C., and Korosteleve, A.A. (2019) Extensive ribosome and RF2 rearrangements during translation termination. eLife 8:e46850. Available at: https://elifesciences.org/articles/46850
- Sauert, M., Temmel, H., and Moll, I. (2015) Heterogeneity of the translational machinery: Variations on a common theme. Biochimie 114:39-47. Available at: https://www.sciencedirect.com/science/article/pii/S0300908414003952