20.4: CO₂ uptake - Calvin Cycle and C3 organisms
- Page ID
- 15050
The source for the organization and some of the text derives from: Sindayigaya and Longhini. https://www.peoi.org/Courses/Courses...chem/biochem18 CC - https://creativecommons.org/licenses...sa/3.0/deed.en
Introduction
We focused on the light reactions of photosynthesis. Now let's turn our attention to the dark reactions which fix CO2 from the air and reduce it with NADPH produced, along with O2, in the light reactions, to produce carbohydrates. The dark reactions don't just occur in the dark. The term is simply used to differentiate them from the light-driven reactions using PSII and PSI. What is so interesting about plants is they produce fuel from CO2 using photons as a source of energy (they are autotrophs) and also consume the fuels they make, using both anaerobic and aerobic respiration pathways. Their biosynthetic reactions take place mostly in the chloroplast, a type of plastid, which are subcellular organelles with specific functions such as photosynthesis or metabolite synthesis and storage. Plants also can not move to acquire fuel and nutrient molecules. They are subject to a large range of growing conditions (differential light qualities and quantities, temperatures, and rainfall levels). Also, plant cells have cell walls in addition to a cell membrane. A simple cartoon showing the major motifs of photosynthesis is shown in Figure \(\PageIndex{1}\).
In this section, we will discuss how CO2 from the atmosphere is "fixed" or "captured" in the formation of the simplest sugars (3 carbon molecules like glyceraldehyde-3-phosphate) in a process called the C3 or Calvin Cycle, which is also called the Calvin–Benson–Bassham (CBB) cycle, or the reductive pentose phosphate cycle (RPP cycle). Plants that use the C3 cycle are logically called C3 plants There are two other major types of carbon capture pathways, the C4 and CAM pathways, which we discuss in the next section. All use a key enzyme, ribulose 1,5-bisphosphate carboxylase (RuBisCo), to covalently fix CO2 into small carbohydrates, 3-phosphoglycerate. RuBisCo is the most abundant protein in the biosphere. Recent estimates suggest that there are about 0.7 gigatons (Gt = 1012 tons) of it, with over 90% in the leaves (about 3% of their weight) of terrestrial plants. It captures about 120 Gt of atmospheric CO2 each year. This enzyme has a second competing enzymatic activity. It is also an oxygenase, which adds to its complexity. That activity captures about 100 Gt of atmospheric O2 each year. In this chapter section, we will give an overview of the C3 pathway and given the importance of RuBisCo, we will focus on it predominately.
Along RuBisCo, plants have pathways to take the fixed CO2 to 3C sugars and then a unique pentose pathway which runs in a reductive fashion to ultimately produce the sugar-containing molecules in plants we are most familiar with, sucrose and the glucose polymer starch.
Plastids
There are several types of these organelles. Photosynthesis occurs in chloroplast which has its own genome, like the mitochondria. Another common type is the amyloplasts, which lack pigmented molecules (i.e. they are colorless) and have no inner membrane. In plants, they are filled with starch. Chloroplasts and amyloplasts can interconvert. Chloroplasts are abundant in green leaves while amyloplasts are predominately found in locations like potato tubers where starch is stored. Light can drive the interconversion of plastids as shown in Figure \(\PageIndex{2}\).
The characteristics and plastid interconversion pathways of the plastids are shown by arrows. The transition to a chloroplast is called “Greening” and is identified with the number “1”. This is mainly triggered by light signals from proplastids, etioplasts, leucoplasts, and chromoplasts. Etioplasts can develop from proplastids in dark conditions and this is identified by the number “2”. The number “3” indicates leucoplast development that is triggered by diverse development processes to generate starch, lipid, and protein-enriched sub-types called amyloplasts, elaioplasts, and proteinoplasts, respectively. Mainly during the ripening stage, diverse types of carotenoid crystals were generated within the plastids called chromoplasts from the proplastids, leucoplasts, and chloroplasts and this is identified with the number “4”. Together with etioplast and leucoplast development (2,3), chromoplast development (4) was identified as a “Non-greening” plastid transition. The loss of green color from the chloroplasts is called “De-greening” and is identified with the number “5”, and these chloroplasts are then transited into leucoplast or gerontoplast by developmental regulation or during senescence, respectively.
CO2 capture and the C3 Cycle
There are in the synthesis of the simplest carbohydrates (3 carbon polyhydroxy- aldehydes and ketones:
- Carbon capture or fixation phase. We prefer the term carbon capture as this term is now used to describe how the world is seeking new ways (other than planting billions of trees) to "capture" excess CO2 emitted through the use of fossil fuels. In a reaction catalyzed by RuBisCo, atmospheric CO2 ultimately react with a 5-carbon acceptor molecule, ribulose 1,5-bisphosphate (Ru1,5-BP, 6 carbons in total), to form two molecules of 3-phosphoglycerate (3PG). (2, 3C molecules).
- Reduction phase: 3-phosphoglycerate is reduced to glyceraldehyde-3-phosphate (G3P). Three CO2s are captured on reaction with 3 Ru1,5-BP to form 6 glyceraldehyde-3-phosphates (G3P). These can readily interconvert to the keto form, dihydroxyacetone phosphate (DHAP).
- Regeneration phase: Five of the six G3Ps (15 Cs) react to form 3 three molecules of ribulose 1,5-bisphosphate (15 C2) to allow the catalytic C3 cycle to continue. The other G3P moves into the stroma, in the form of DHAP where it can be used in gluconeogenesis (reductive biosynthesis) of glucose. This can be converted to polymer starch and also the disaccharide sucrose (which we will discuss in a future session).
An overview of the Calvin or C3 cycle is shown below in Figure \(\PageIndex{3}\).
The stoichiometry can be confusing until you count the actual number of carbon atoms and realize that the cycle has to run 3 times to enable 3 carbon atoms from 3 CO2 molecules to produce one net glyceraldehyde-3-phosphate (G3P). The G3P leaves the C3 cycle at the low left for glucose synthesis. The conversion of the 5 G3Ps that reform Ru1,5-BP requires ATP as shown below:
\[\ce{5 glyceraldehyde-3P + 3 ATP → 3 ribulose-1,5-2P + 3 ADP + 2 P_i} \nonumber \]
with \(\ce{P_i}\) indicating inorganic phosphate. Hence the net equation for 3 turns of the cycle, sufficient to produce 1, G3P is:
\[\ce{3 CO2 + 6 NADPH + 6 H^{+} + 9 ATP + 5 H2O → glyceraldehyde-3-phosphate (G3P) + 6 NADP^{+} + 9 ADP + 8 P_i } \nonumber \]
Even though glucose is not a product of the Calvin cycle, some texts use the following equation to show the stoichiometry to run the C3 cycle enough times (6) to fix 6 \(\ce{CO2}\) molecules, enough to make 1 glucose from a simple carbon atom counting perspective.
\[\ce{6 CO2 + 12 NADPH + 12 H^{+} + 18 ATP + 10 H2O → 2 glyceraldehyde-3-phosphate (G3P) + 12 NADP^{+} + 18 ADP + 16 P_i } \nonumber \]
Remember that NADPH and ATP are produced in the light reactions in about the same ratio as they are used in the C3 cycle (2NADPH/3ATPs). The net 8 Pis made as products will react with 8 ADP to regenerate 8 ATP in the light reaction. The 9th Pi is incorporated in a triose-phosphate in the light reaction, so one Pi must be imported from the cytoplasm by an inner membrane triose-phosphate/phosphate translocator, which we will discuss below. In the dark, when ATP and NADPH are not produced, CO2 capture also is inhibited.
A more detailed diagram showing the detailed reactions to regenerate ribulose 1,5-bisphosphate (Ru1,5BP) is shown in Figure \(\PageIndex{4}\).
Abbreviated reactions for the synthesis of sucrose, glucogenic amino acids, and fatty acids are also shown.
You should note the reactions for the conversion of the 6C sugar molecule fructose-6-P (F6P) (glycolytic and gluconeogenic intermediate) to the 5C molecule Ru5P and Ru1-5BP, are analogous to the reactions of the nonoxidative part of the pentose phosphate pathway (PPP) pathway which generates 5C sugars for the synthesis of nucleotides, nucleic acids, and some amino acids. Hence we won't discuss them further.
Carbon capture of CO2 into 3-Phosphoglycerate - RuBisCo
This key enzyme requires a Mg2+ ion and proceeds through a carbamoylated lysine side chain. The Mg2+ ion orients key side chains. The resulting 6C molecule cleaves into two 2 molecules of 3PG.
The RuBisCo family of enzymes can adopt different quaternary structures. A homodimer of two large subunits is the minimum catalytic structure. Often there are in addition two small subunits. The most common form is a 16mer which is found in cyanobacteria, red and brown algae, and all higher plants.
A possible mechanism of RuBisCo from Synechococcus elongatus, a unicellular cyanobacterium, is shown in Figure \(\PageIndex{5}\).
The carbamoylated lysine side chain is shown in green. Ribulose 1,5- bisphosphate is converted to an enediolate which engages in a nucleophilic attack on the CO2 to form a 6C sugar. Hydroxylation at C-3 of this sugar is followed by aldol cleavage. Ultimately two 3PGs are produced, one of which contains the carbon atom from CO2 (red).
Figure \(\PageIndex{6}\) shows an interactive iCn3D model of a single heavy and light chain of ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCo) from Synechococcus PCC6301 (1RBL). (long load time)
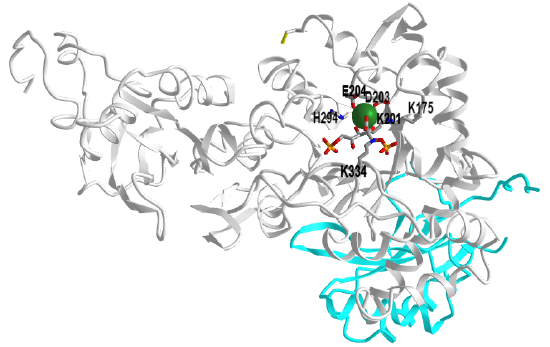
The light chain is shown in cyan and key residues in the heavy chain are shown in CPK-colored sticks and labeled. Bound to the heavy chain is a substrate analog/inhibitor, 2-carboxyarabinitol-1,5-diphosphate. It is produced in plants and in the dark, it inhibits the enzyme. With increasing lights, its concentration decreases.
Recent Updates (08/14/23) - Rubisco Reacts with both CO2 and O2
Rubisco is a slow enzyme with a kcat of around 2-10 CO2/sec. In addition, it can bind another substrate, O2, and engage in a competing reaction of photorespiration (oxidation) of ribulose 1,5-bisphosphate to form one molecule of 3-phosphoglycerate (3PG) and one molecule of 2-phosphoglycolate (2PG), as shown (incompletely) in Figure \(\PageIndex{7}\) below.
Figure \(\PageIndex{7}\): RuBP conversion by Rubisco through the carboxylase (a) and the oxygenase (b) reactions. Tommasi, I.C. The Mechanism of Rubisco Catalyzed Carboxylation Reaction: Chemical Aspects Involving Acid-Base Chemistry and Functioning of the Molecular Machine. Catalysts 2021, 11, 813. https://doi.org/10.3390/catal11070813. CC BY) license (https://creativecommons.org/licenses/by/4.0/
Following RuBP (1) enolization, the 2,3-enol(ate) intermediate (2) may react with CO2(a) or O2(b) co-substrates. The carboxylase reaction produces the 2-carboxy-3-keto-arabinitol 1,5-bisphosphate intermediate (3) undergoing protonation to the 2-carboxylic acid before hydration. The C2-C3-scission reaction in C3-gemdiolate (5) is described as occurring in a concerted mechanism upon P1 protonation producing two molecules of 3-phospho-D-glycerate (3PGA, 6). The oxygenase reaction produces 3-phospho-D-glycerate (3PGA,6) and 2-phosphoglycolate (2PG,7)
How does the enzyme differentiate the two nonpolar substrates, CO2 and O2? Given the symmetric arrangement of δ+ and δ- charges in CO2, it has a net 0 dipole, as does O2, so it would not align/orient in a field generated by two poles (+ and -). However, CO2, but not O2, would align in a field generated by four charged poles so it has a quadrupole moment, as shown in Figure \(\PageIndex{8}\) below.
Figure \(\PageIndex{8}\): CO2 aligning in a quadrupole field. (field lines from https://commons.wikimedia.org/wiki/F...quadrupole.svg)
The dipole unit is the debye and the quadrupole unit can be expressed in debye.Angstrom. Table \(\PageIndex{1}\) below shows some values for dipole and quadrupole moments for simple gases. CO2 has the highest quadrupole moment of all these simple gases.
| Molecule | Dipole moment (D) | Quadrupole moment (D Å) |
| CO2 | 0.000 | 4.30 |
| CH4 | 0.000 | 0.02 |
| H2 | 0.000 | 0.66 |
| O2 | 0.000 | 0.39 |
| CO | 0.112 | 2.5 |
| N2 | 0.000 | 1.52 |
Table \(\PageIndex{1}\): Dipole and Quadrupole moments for some simple gases. Castro-Muñoz, R., Fíla, V., 2018. Progress on Incorporating Zeolites in Matrimid®5218 Mixed Matrix Membranes towards Gas Separation. Membranes 8, 30.. https://doi.org/10.3390/membranes8020030
The active site has a high electrostatic field gradient in the dimeric form of the enzyme. Figure \(\PageIndex{9}\) shows an interactive iCn3D model showing the electrostatic potential surface in the active site between two heavy chains of spinach ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCo) (8RUC). It shows that is complex.
Figure \(\PageIndex{9}\): Electrostatic potential surface in the active site between two heavy chains of spinach ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCo) (8RUC). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...2JG3x4csVnEq67
Two heavy chains are shown in light pink and cyan, and one light chain is shown in gray. The active site regions are shown as an electrostatic surface potential with blue positive and red negative. The model has an activated substrate analog, 2-carboxyarabinitol bisphosphate, shown spacefill (hard to see given the electrostatic surface potential). The residue labeled 201KCX is the carbamate of Lys201.
Presumably, the electrostatic field in the active site facilitates through subtle interaction of the quadrupole CO2 compared to O2.
The mechanisms that differentiate the binding of CO2 and O2 must also overcome the high intracellular concentration of O2 (around 250 μM) compared to CO2 (7–8 μM in C3 plants and 80 μM in C4 plants). The enhanced affinity for CO2 (about 30-fold) compared to O2 helps overcomes these concentration barriers. No classic "binding pocket" exists for CO2 and O2 so diffusion and binding is likely guided by the electrostatic potential gradients along the diffusion surface. Figure \(\PageIndex{10}\) below the electrostatic potential molecular surface of O2 and CO2 (top), calculated from electron density measurement using quantum mechanics, and the electrostatic surface of their binding pockets
Figure \(\PageIndex{10}\): Electrostatic potential molecular surface of O2 and CO2 (top) and the electrostatic surface of their binding pockets. Tommasi, I.C. et al., ibid.
(top) Computed electrostatic potential molecular surfaces of CO2 (left) and O2 (right). The color scheme follows commonly accepted conventions: blue, positive; red, negative. The value of the Qzz component of the quadrupole moment, as calculated by Stec, is −3.239 e a02 for CO2 and −0.232 e a02 for O2. Note that the ratio of these values is about 14, about equal to the earlier quadrupole moments discussed above with units of debeye.Å)
(bottom) a ribbon representation of the catalytic domain with bound gaseous ligands and surfaces colored by the electrostatic potential. O2 (in red) and CO2 (in purple) lie in a positively charged cavity (blue) of the TIM barrel. (Figure from ref. [15], used by permission of PNAS (copyright © 2012)).
Both CO2 and O2 are situated in a tiny "cavity" that is blue (positive potential, C-terminal domain) and just above it red (negative potential, N-terminal domain). The quadrupole moment of CO2 is 10-15x that of O2 which helps explain its higher affinity in the localized electrostatic potential gradients around the gas molecules.
Molecular dynamic (MD) simulations show an interaction preference for CO2. There are many subunit-subunit interfaces and all appear in MD to preferentially interact with CO2, which probably moves from the solvent through large:small subunit interface to the active site. The CO2 does not localize long at any residues but seems to occupy areas instead. Since CO2 has no dipole, it locates more closely to small hydrophobic side chain (Ala, Val, Leu, Ile) and the main chain. CO2 has more interaction in every active site as well as the large:large subunit interface (whose electrostatic potential in the active site is shown above) and in the large:small subunit interface.
In efforts to quantitate the preference of CO2 over O2 using MD, investigators found that over many different species of rubisco, the relative distribution of CO2 and O2 to the small and large was on average 1.8 for CO2 and 1.4 for O2 with the number of oxygen bound to either subunit lower. This is true even though CO2 has a lower solvation energy than O2 in water, so additional energy must be spent to differentially desolvate CO2. The hydrophobic interactions likely promote the movement of CO2 to the active site where electrostatic-based potentials likely favor CO2 binding. These results suggest that the small subunit acts like a "miniresevoir" for O2 which then diffuse to the large subunit region of the active site. From a simple thermodynamic perspective, CO2 would be favored to move along the surface and through spaces in the protein guided by transient interactions that be water. The active site in rubisco is not in a deep pocket but rather in shallow groves near the surface. Although the enzyme is slow (2-10 CO2/s), it's not much slower than the average enzyme. The median turnover number kcat (under saturating conditions) of enzymes is about 10 s-1 with most following between 1-100. Its concentration is very high in chloroplasts, which helps increase the fixing of CO2.
The next step: 3-Phosphoglycerate to Glyceraldehyde 3-Phosphate and dihydroxyacetone phosphate
The 3PG produced by RuBisCo is converted to the triose glyceraldehyde-3-P (G3P) (which can readily isomerize to dihydroxyacetone phosphate) using typical glycolytic enzymes run in reverse except that NADPH is used as a reductant instead of NADH. In addition, the stromal and cytosolic enzymes derive from different genes. The remaining G3P not used to resynthesize ribulose 1,5BP can be used for the synthesis of starch, sucrose, etc, as illustrated in Figure 4 above.
Exchange of trioses and phosphate across the inner membrane
The inner chloroplast membrane has a triose-phosphate/phosphate translocator (TPT), an antiporter that brings into the stroma Pi in exchange for a triose phosphate, either dihydroxyacetone phosphate or 3-phosphoglycerate. The importance of this was discussed above. The exported triose can be used for the synthesis of sucrose, which can be transported around the plant as a source of carbon. Trioses within the chloroplast can also be converted to glucose and onto glycogen as the organelle becomes an amyloplast. If the translocator is inhibited, Pi would decrease in the chloroplast, which would decrease ATP and also starch synthesis.
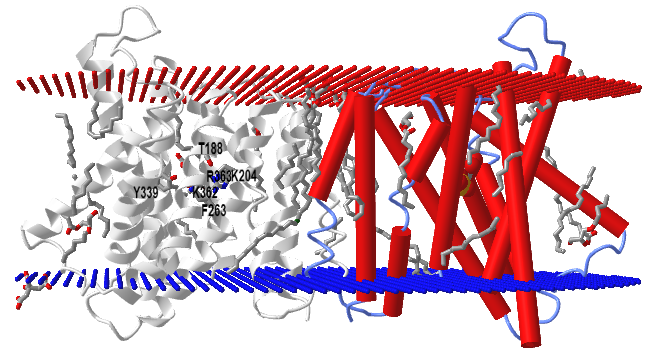
The structure of TPT has been determined with the bound ligands, 3-phosphoglycerate and inorganic phosphate, in an occluded conformation from Galdieria sulphuraria, an extremophilic unicellular species of red algae. Figure \(\PageIndex{11}\) shows an interactive iCn3D model of a triose-phosphate/phosphate translocator from the red algae (5Y78).
.png?revision=1&size=bestfit&width=455&height=245)
The model shows two monomers, one with red cylindrical alpha helices and spacefill 3-phosphoglycerate. The other subunit is shown in gray with the 3PG in colored sticks and conserved residues (T188, K204, F263, Y339, K362, R363) that make interacts with both Pi and 3PG. There would presumably be an outward- and inward-open conformation that is triggered on ligand binding.
Activity Regulation by Light
Given their role in photosynthesis, you would expect even the dark reaction enzymes would be regulated by light. Indeed, four C3 cycle enzymes are. They are ribulose 5-phosphate kinase, fructose 1,6-bisphosphatase, sedoheptulose 1,7-bisphosphatase, and glyceraldehyde 3-phosphate dehydrogenase. The regulation is affected by photon-induced disulfide bond formation between two cysteine side chains in the enzymes. When oxidized (disulfide bond form), the enzymes are inactive. Under light conditions, PSII, cyto b6f, and PSI work in electron transport to move electrons from H2O to ferredoxin and onto a small soluble protein thioredoxin which has a disulfide. The enzyme catalyzing this last step is ferredoxin-thioredoxin reductase. On reduction, the disulfide in thioredoxin is cleaved, and the now free sulfhydryls in thioredoxin are used to cleave the disulfide in the 4 enzymes mention above, in a similar fashion to how β-mercaptoethanol in excess can cleave disulfides in proteins. This leads to conformational changes in the four enzymes which activate them. In the absence of light, the process reverses, and the enzymes are inhibited. For fuel at night, plants mobilize starch for energy.
A simple mechanism to show how thioredoxin catalyzes disulfide bond reduction in target proteins is shown in Figure \(\PageIndex{12}\).
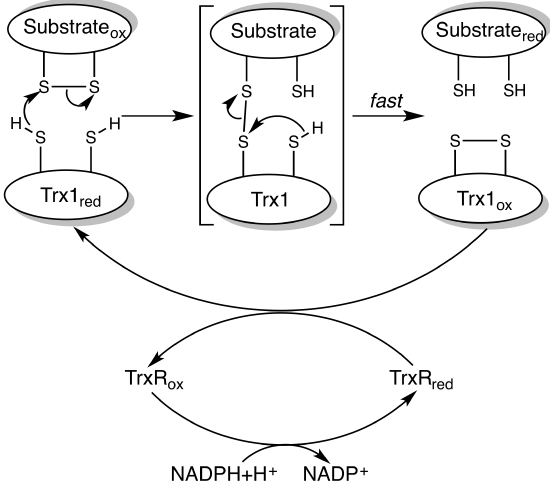
The first enzyme in the oxidative branch of the pentose pathway, glucose 6-phosphate dehydrogenase, uses NADP+ as an oxidizing agent, producing NADPH. In the light, there is lots of NADPH produced from the light reactions of photosynthesis so it makes biological sense that under these conditions, glucose 6-phosphate dehydrogenase activity is inhibited. It is so, also by the cleavage of a critical disulfide bond, but in this case, it results in enzyme inactivation.
We saw the role of thioredoxin in the previous chapter section when we discussed the regulation of photosynthesis as well as the ATP synthase of the chloroplast.
Figure \(\PageIndex{13}\) shows an interactive iCn3D model showing a comparison of the structures of oxidized (1ERU) and reduced (1ERT) human thioredoxin.
Figure \(\PageIndex{13}\): Comparison of the structures of oxidized (1ERU) and reduced (1ERT) human thioredoxin. (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...XPVsErV2JWxdA6.
The two subunits of thioredoxin, linked by a disulfide are shown in gray. Press the "a" key to toggle between the oxidized form, with Cys32-Cys35 disulfide shown as a yellow stick, and the reduced form with the reduced and separated Cys 32 and Cys 35 shown in colored spheres. Not that the hydrogen covalently attached to the free cysteine side chain does not show in a crystal PDB structure.
SUMMARY
Photosynthesis in vascular plants takes place in chloroplasts. In the CO2-assimilating reactions (the Calvin cycle), ATP and NADPH are used to reduce CO2 to triose phosphates. These reactions occur in three stages: the fixation reaction itself, catalyzed by Rubisco; reduction of the resulting 3-phosphoglycerate to glyceraldehyde 3-phosphate; and regeneration of ribulose 1,5-bisphosphate from triose phosphates. Rubisco condenses CO2 with ribulose 1,5-bisphosphate, forming an unstable hexose bisphosphate that splits into two molecules of 3-phosphoglycerate. Rubisco is activated by covalent modification (carbamoylation of Lys201) catalyzed by Rubisco activase and is inhibited by a natural transition-state analog, whose concentration rises in the dark and falls during daylight. Stromal isozymes of the glycolytic enzymes catalyze the reduction of 3-phosphoglycerate to glyceraldehyde 3-phosphate; each molecule reduced requires one ATP and one NADPH. The cost of fixing three CO2 into one triose phosphate is nine ATP and six NADPH, which are provided by the light-dependent reactions of photosynthesis. An antiporter in the inner chloroplast membrane exchanges Pi in the cytosol for 3-phosphoglycerate or dihydroxyacetone phosphate produced by CO2 assimilation in the stroma. Oxidation of dihydroxyacetone phosphate in the cytosol generates ATP and NADH, thus moving ATP and reducing equivalents from the chloroplast to the cytosol. Four enzymes of the Calvin cycle are activated indirectly by light and are inactive in the dark so that hexose synthesis does not compete with glycolysis—which is required to provide energy in the dark.
Photorespiration - RuBisCo/Oxygenase and the Glycolate Cycle
As autotrophs, plants make their fuels. They use that fuel to make ATP to power endergonic reactions like protein synthesis, cell division, etc. As eukaryotic cells, they have mitochondria and can use both aerobic and anaerobic respiration to produce ATP. In the dark, when photons are not present, they carry out mitochondrial aerobic respiration as they break down carbohydrates to CO2 and water, the reverse of photosynthesis.
They also use O2 in another process that is driven by light. The same enzyme that captures carbon, RuBisCo, has oxygenase activity. RuBisCo uses O2 in a process called photorespiration, which produces CO2 in a competing reaction. Same enzyme, different substrates! The final products of the reaction with CO2 using RuBisCo are two 3C molecules, 3-phosphoglycerate (3PG). Using O2 as a substrate produces 1 molecule of the 3C 3PG and 1 molecule of a 2C analog, 2-phosphoglycolate (not 2-phosphoglycerate). 2-phosphoglycolate is also named carboxymethylphosphate. About one out of every four turnovers of the enzyme produced this metabolic dead product. Given this non-trivial side reaction, the enzyme should be called ribulose 1,5-bisphosphate carboxylase/oxygenase.
You may ask why would such a critical enzyme evolved to a form which is quite inefficient. One explanation is that the enzyme "finished" its evolution before the great oxygenation event when dioxygen rose to the levels we see now (20%). Before that, little oxygen was available to compete with the trace gas CO2, which is around 0.04% of the atmosphere. (Even though CO2 is considered a trace gas, its present concentration is around 420 parts per million (ppm), levels which are warming our planet and which have not been seen for 3 million years, when Arctic forests and camels were present).
Compare the KM values (9 μM for CO2 and 350 μM for O2 or 39x higher for O2) for the enzyme and the equilibrium concentrations of the gases in aqueous solution (11 μM for CO2 and 250 μM for O2 or 23x higher for O2). The higher KM for O2 is nearly offset by O2's greater solubility so modern conditions lead to a significant waste of the CO2 capture efficiency of RuBisCo/Oxy. At higher temperatures in a warming world, the equilibrium ratio of solution concentrations of O2/CO2 increases as does the affinity (based crudely on KM values) of CO2. Both of these exacerbate the wasteful oxygenase activity effect. Finally, as CO2 is captured by the enzyme, the ratio of the local concentrations of O2/CO2 also goes up. All of these make the efficiency of RuBisCo worse.
Figure \(\PageIndex{14}\) shows a mechanism for the reaction of both CO2 and O2 with RuBisCo/Oxygenase.
Note that in contrast to most oxygenases, no cofactor is required for the RuBisCo/Oxygenase.
The glycolate pathway
The 2-phosphoglycolate (carboxylmethyl phosphate) "waste" product of the oxygenase activity of RuBisCo/Oxygenase is recycled through a complex pathway that is called "photorespiration". It occurs in three different organelles, the chloroplast, the peroxisome, and the mitochondria. Part of the generalized pathway is shown in Figure \(\PageIndex{15}\).
Multiplying the stoichiometry represented in the figure by 2 shows that 2 molecules of 2-phosphoglycolate produce 2 molecules of glycine. These get converted to two molecules of serine. We will see the mechanisms for some of these reaction in the chapter on amino acid metabolism. The net reaction is:
\[\ce{2 Glycine + NAD^{+}+ H2O → serine + CO2 + NH3 + NADH} \nonumber \]
The serine is eventually converted to 3-phosphoglycerate, which can be used again in the C3 cycle. Note that CO2 is produced in the glycolate pathways that started with the use of O2 as a substrate by RuBisCo/Oxygenase. Hence the whole system uses O2 and produces one CO2 so the combined reactions are usually called photorespiration. It's not an ideal term since it is wasteful, compared to mitochondrial respiration. Some prefer to call the combined pathway of Rubisco oxygenase and the glycolate pathways the C2 cycle.




.png?revision=1&size=bestfit&width=399&height=269)
_and_reduced_(1ERT)_human_thioredoxin.png?revision=1&size=bestfit&width=386&height=233)