25.1: DNA-Dependent Synthesis of RNA
- Page ID
- 15197
Types of RNA
Structurally speaking, ribonucleic acid (RNA), is quite similar to DNA. However, whereas DNA molecules are typically long and double-stranded, RNA molecules are much shorter and are typically single-stranded. A ribonucleotide within the RNA chain contains ribose (the pentose sugar), one of the four nitrogenous bases (A, U, G, and C), and a phosphate group. The subtle structural difference between the sugars gives DNA added stability, making DNA more suitable for the storage of genetic information, whereas the relative instability of RNA makes it more suitable for its more short-term functions. The RNA-specific pyrimidine uracil forms a complementary base pair with adenine and is used instead of the thymine that is found in DNA. Even though RNA is single-stranded, most types of RNA molecules show extensive intramolecular base pairing between complementary sequences within the RNA strand, creating a predictable three-dimensional structure essential for their function, as shown in Figure \(\PageIndex{1}\) and Figure \(\PageIndex{2}\).
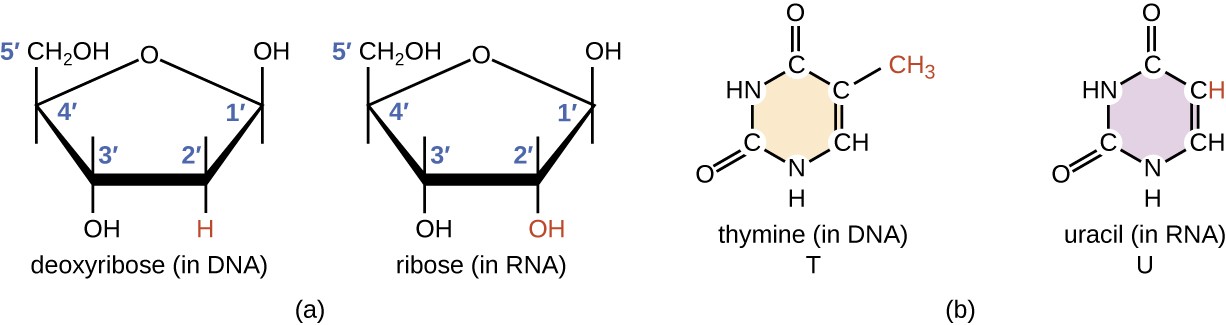
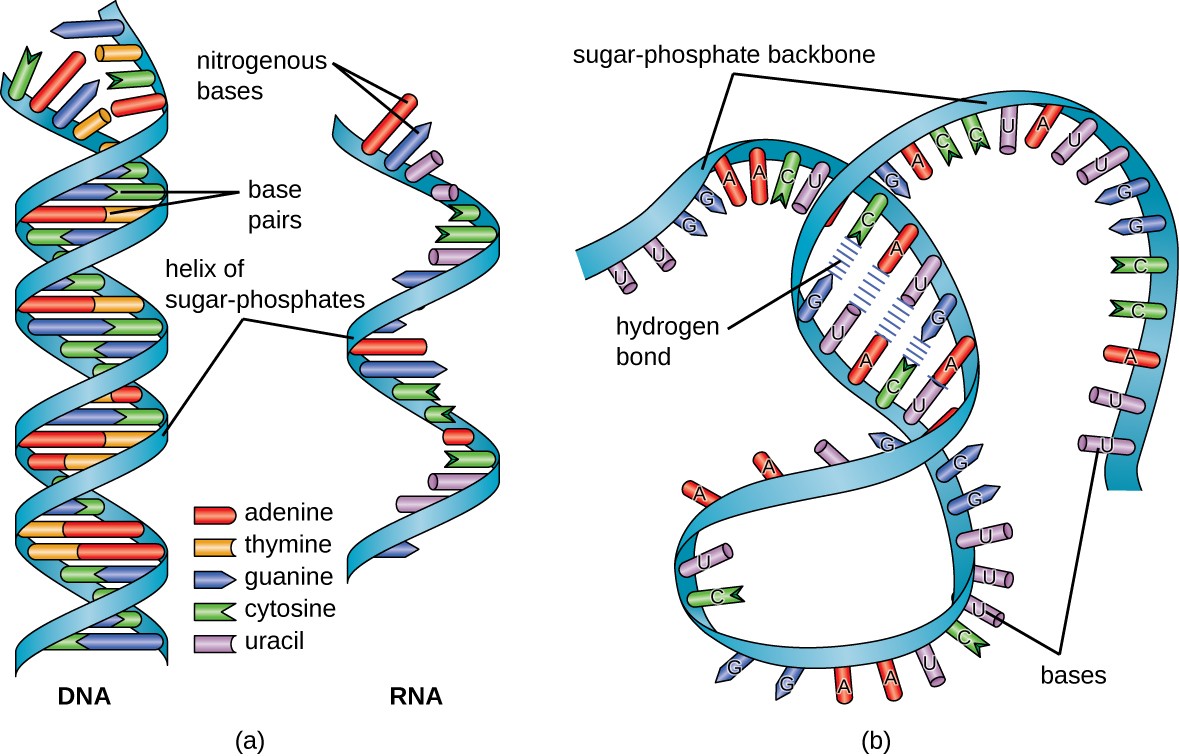
RNA can largely be divided into two types, one that carries the code for making proteins or coding RNA, which is also called messenger RNA (mRNA), and non-coding RNA (ncRNA). The ncRNA can be subdivided into several different types, depending either on the length of the RNA or on the function. Size classification begins with the short ncRNAs (~20–30 nt), which include microRNAs (miRs), and small interfering (siRNAs); the small ncRNAs up to 200 nt, which include transfer RNA (tRNA), small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA); and long ncRNAs ( > 200 nt), which include ribosomal RNA (rRNA), enhancer RNA (eRNA) and long intergenic ncRNAs (lincRNAs), among others.
Cells access the information stored in DNA by creating RNA, through the process of transcription, which then directs the synthesis of proteins through the process of translation. The three main types of RNA directly involved in protein synthesis are messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). The mRNA carries the message from the DNA, which controls all of the cellular activities in a cell. If a cell requires a certain protein to be synthesized, the gene for this product is “turned on” and the mRNA is synthesized through the process of transcription. The mRNA then interacts with ribosomes and other cellular machinery to direct the synthesis of the protein it encodes during the process of translation. mRNA is relatively unstable and short-lived in the cell, especially in prokaryotic cells, ensuring that proteins are only made when needed.
rRNA and tRNA are stable types of RNA. In prokaryotes and eukaryotes, tRNA and rRNA are encoded by the DNA, where they are transcribed into long RNA molecules that are subsequently cut to release smaller fragments containing the individual mature RNA species. In eukaryotes, synthesis, cutting, and assembly of rRNA into ribosomes takes place in the nucleolus region of the nucleus, but these activities occur in the cytoplasm of prokaryotes. Within the nucleolus region, ribosome assembly requires the activity of numerous snoRNAs.
Ribosomes are composed of rRNA and protein. As its name suggests, rRNA is a major constituent of ribosomes, composing up to about 60% of the ribosome by mass and providing the location where the mRNA binds. The rRNA ensures the proper alignment of the mRNA, tRNA, and the ribosomes; the rRNA of the ribosome also has an enzymatic activity (peptidyl transferase) and catalyzes the formation of the peptide bonds between two aligned amino acids during protein synthesis (Figure 26.1.3). Although rRNA had long been thought to serve primarily a structural role, its catalytic role within the ribosome was shown in 2000. Scientists in the laboratories of Thomas Steitz (1940–) and Peter Moore (1939–) at Yale University were able to crystallize the ribosome structure from Haloarcula marismortui, a halophilic archaeon isolated from the Dead Sea. Because of the importance of this work, Steitz shared the 2009 Nobel Prize in Chemistry with other scientists who made significant contributions to the understanding of ribosome structure. The structure and function of ribosomes will be discussed in further detail in Chapter 27.
Transfer RNA (tRNA) is the third prominent type of RNA involved in protein translation. tRNAs are usually only 70–90 nucleotides long. They carry the correct amino acid to the site of protein synthesis in the ribosome. It is the base pairing between the tRNA and mRNA that allows for the correct amino acid to be inserted in the polypeptide chain being synthesized, as shown in Figure \(\PageIndex{3}\). Any mutations in the tRNA or rRNA can result in global problems for the cell because both are necessary for proper protein synthesis.
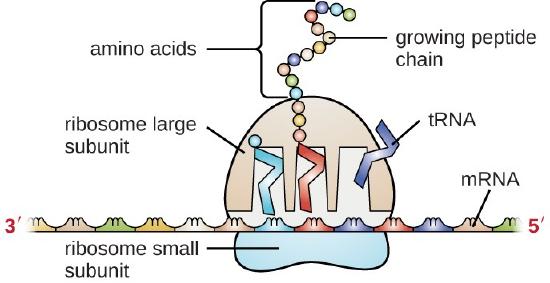
As described previously, some RNA molecules have enzymatic properties and serve as ribozymes. Within this chapter, the activity of snRNAs during the process of intron removal from mRNA sequences function as ribozymes and will be described. Furthermore, a detailed description of the enzymatic features of the ribosome structure will be provided in Chapter 27.
Other small ncRNAs and lncRNA molecules play a role in the regulation of transcriptional and translational processes. For example, the post-transcriptional expression levels of many genes can be controlled by RNA interference, in which miRNAs, specific short RNA molecules, pair with mRNA regions and target them for degradation, as shown in Figure \(\PageIndex{4}\). This process is aided by protein chaperones called argonautes. This antisense-based process involves steps that first process the miRNA so that it can base-pair with a region of its target mRNAs. Once the base pairing occurs, other proteins direct the mRNA to be destroyed by nucleases. Fire and Mello were awarded the 2006 Nobel Prize in Physiology or Medicine for this discovery.
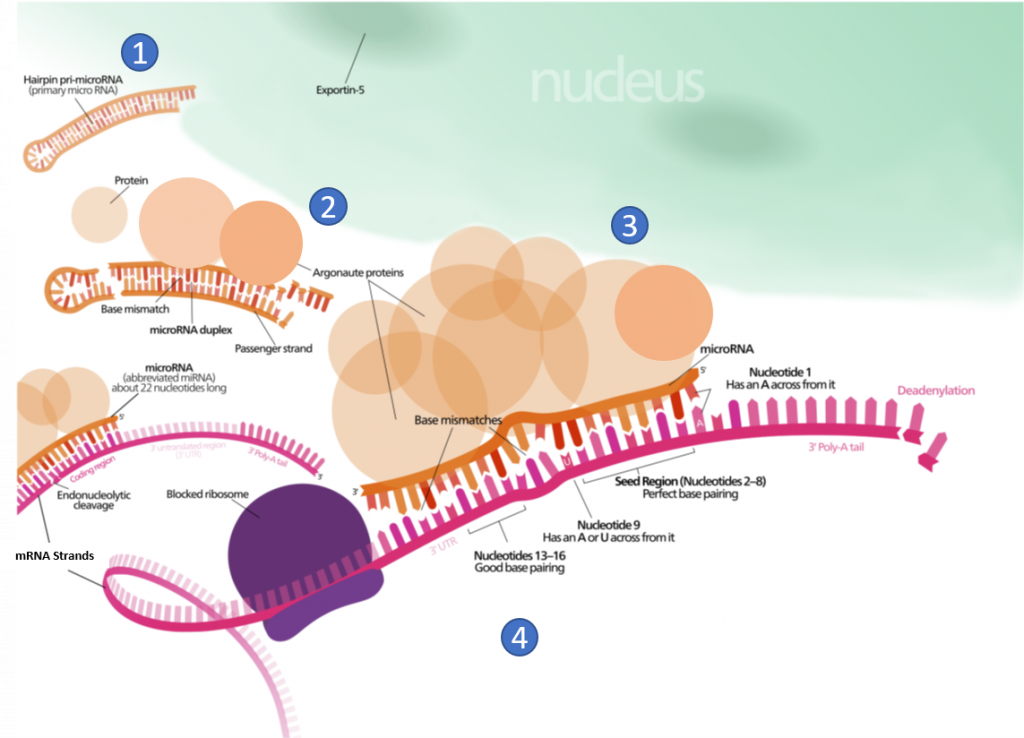
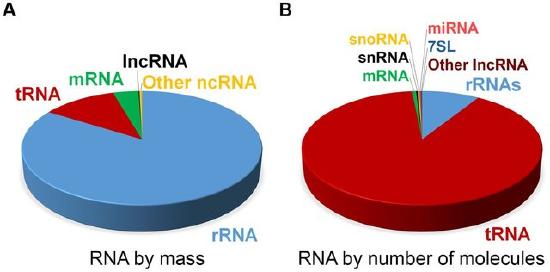
At steady state, the vast majority of human cellular RNA consists of rRNA (∼90% of total RNA for most cells) as shown in Figure \(\PageIndex{5}\). Although there is less tRNA by mass, their small size results in their molar level being higher than rRNA (Figure 26.1.5). Other abundant RNAs, such as mRNA, snRNA, and snoRNAs are present in aggregate at levels that are about 1–2 orders of magnitude lower than rRNA and tRNA (Figure 26.1.5). Certain small RNAs, such as miRNA and piRNAs can be present at very high levels; however, this appears to be cell type dependent. lncRNAs are present at levels that are two orders of magnitude less than total mRNA. Although the estimated number of different types of human lncRNAs may have a very restricted expression pattern and thus, accumulate to higher levels within specific cell types. For example, sequencing of mammalian transcriptomes has revealed more than 100,000 different lncRNA molecules can be produced, compared with the approximate 20,000 protein-coding genes. The diversity and functions of the transcriptome within biological processes are currently a highly active area of research.
RNA Polymerases
This chapter will focus on the synthesis of RNA by DNA-dependent RNA Polymerase Enzymes (RNAPs). These enzymes are required to carry out the process of transcription and are found in all cells ranging from bacteria to humans. All RNAPs are multi-subunit assemblies, with bacteria having five core subunits, α2ββ'ω, that have homologs in archaeal and eukaryotic RNAPs. Bacterial RNAPs are the simplest form of RNA polymerases and provide an excellent system to study how they control transcription.
Prokaryotic RNA Polymerase Enzymes
The RNAP catalytic core within bacteria contains five major subunits (α2ββ'ω) (see Figure \(\PageIndex{7}\)) below. To position this catalytic core onto the correct promoter requires the association of a sixth subunit called the sigma factor (σ). Within bacteria, there are multiple different sigma factors that can associate with the catalytic core of RNAP that help to direct the catalytic core to the correct DNA locations, where RNAP can then initiate transcription. For example, within E. coli σ70 is the housekeeping sigma factor that is responsible for transcribing most genes in growing cells. It keeps essential genes and pathways operating. Other sigma factors are activated during certain environmental situations, such as σ38 which is activated during starvation or when cells reach the stationary phase. When the sigma subunit associates with the RNAP catalytic core, the RNAP has then formed the holoenzyme. When bound to DNA, the holoenzyme conformation of RNAP can initiate transcription.
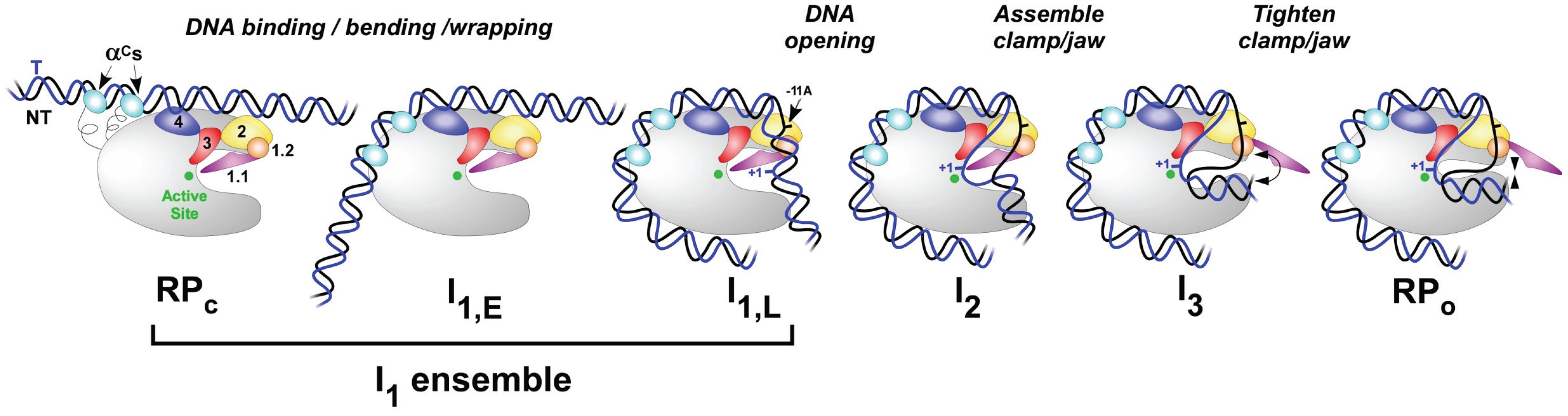
Transcription takes place in several stages. To start with, the RNA polymerase holoenzyme locates and binds to promoter DNA. At this stage the RNAP holoenzyme is it the closed conformation (RPc), as shown in Figure \(\PageIndex{6}\). Initial specific binding to the promoter by sigma factors of the holoenzyme sets in motion conformational changes in which the RNAP molecular machine bends and wraps the DNA with mobile regions of RNAP playing key roles, as shown in Figure \(\PageIndex{6}\). Next, RNAP separates the two strands of DNA and exposes a portion of the template strand. At this point, the DNA and the holoenzyme are said to be in an ‘open promoter complex’ (RPo), and the section of promoter DNA that is within it is known as a ‘transcription bubble’. Intermediates(I1-3) between RPc and RPo occur.
In bacterial systems, the sigma factor locates the transcriptional start site using key DNA sequence elements located at -35 nucleotides and -10 nucleotides from the transcriptional initiation site, as shown in Panel A of Figure \(\PageIndex{7}\). This region is called the Pribnow box. For RNAP from Thermus aquaticus, the −35 element interacts exclusively with
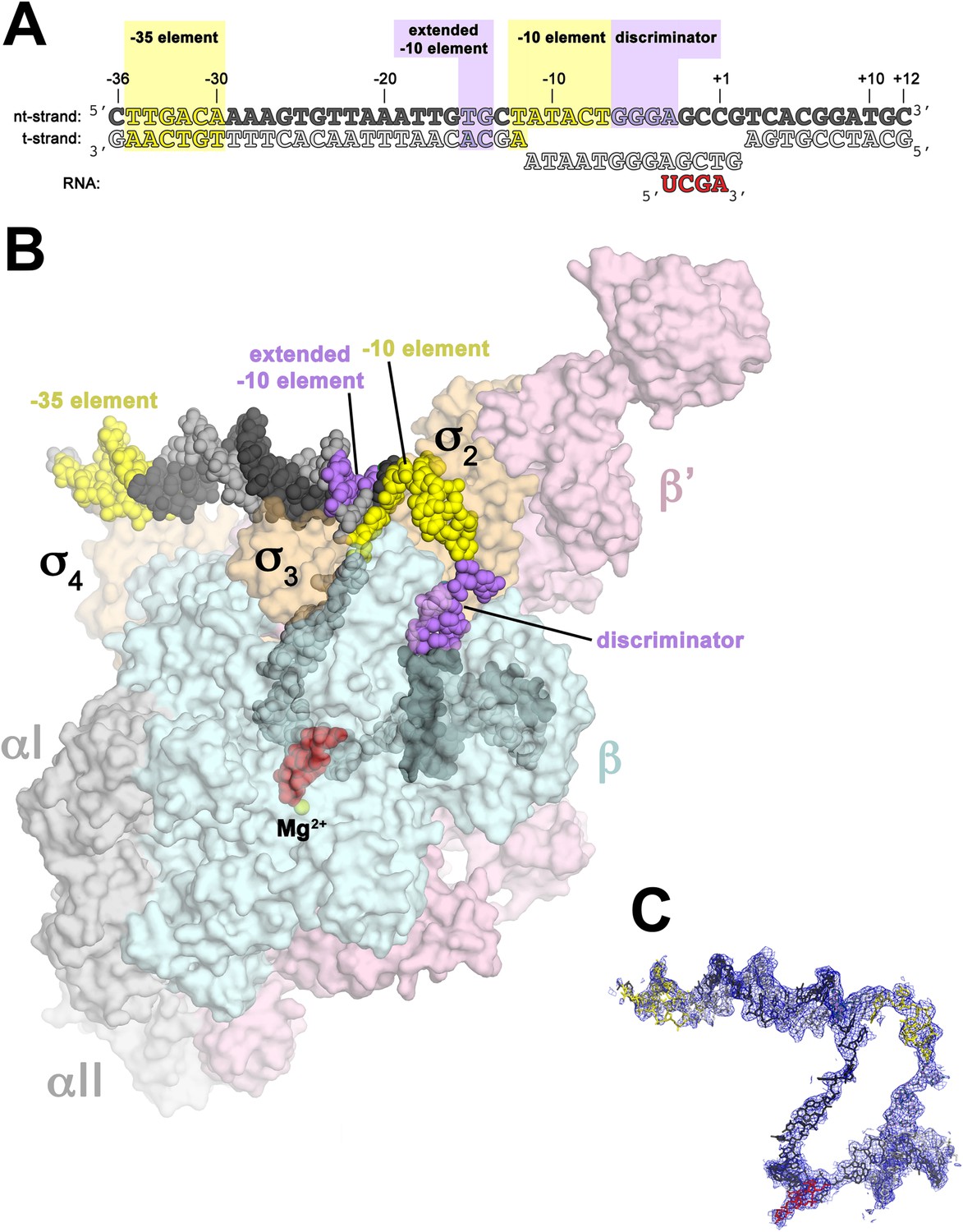
Panel (A) shows oligonucleotides used for the crystallization of the RNAP holoenzyme in the open conformation. The numbers above denote the DNA position with respect to the transcription start site (+1). The −35 and −10 (Pribnow box) elements are shaded yellow, and the extended −10 and discriminator elements are purple. The nontemplate-strand DNA (top strand) is colored dark grey; template-strand DNA (bottom strand), light grey; RNA transcript, red.
Panel (B) shows the overall structure of RNAP holoenzyme in the open conformation bound with the DNA nucleotides. The nucleic acids are shown as CPK spheres and color-coded as in diagram A. Within RNAP, the αI, αII, ω, are shown in grey; β in light cyan; β′ in light pink; Δ1.1σA in light orange. The Taq EΔ1.1σA (Taq derives from Thermus aquaticus) is shown as a molecular surface and the forward portion of the RNAP holoenzyme is transparent to reveal the RNAP active site Mg2+ (yellow sphere) and the nucleic acids held inside the RNAP active site channel.
Panel (C) Electron density and model for RNAP holoenzyme nucleic acids in the open conformation. Color coding matches diagram A.
Figure \(\PageIndex{8}\) shows an interactive iCn3D model of the T. aquaticus transcription initiation complex containing bubble promoter and RNA (4XLN).
It is colored coded in fashion similar to that shown in Figure \(\PageIndex{7}\).
The rendering of the DNA is as follows:
- The non-template (nt-strand) DNA is colored dark grey; spheres
- template (t-strand) DNA, light grey; spheres
- Pribnow box yellow
- discriminator purple
The proteins are shown as surfaces with transparent secondary structures underneath. The color is as follows:
- (αI, αII, ω, light yellow;
- β, light cyan;
- β′, light pink;
- Taq EΔ1.1σA, light orange),
The RNA polymerase active site is located at the Mg2+ (black sphere) binding site. The nucleic acids are inside the RNAP active site channel.
Eukaryotic RNA Polymerase Enzymes
In eukaryotic cells, three RNAPs (I, II, and III) share the task of transcription, the first step in gene expression. RNA Polymerase I (Pol I) is responsible for the synthesis of the majority of rRNA transcripts, whereas RNA Polymerase III (Pol III) produces short, structured RNAs such as tRNAs and 5S rRNA. RNA Polymerase II (Pol II) produces all mRNAs and most regulatory and untranslated RNAs.
The three eukaryotic RNA polymerases contain homologs to the five core subunits found in prokaryotic RNAPs. In addition, the eukaryotic Pol I, Pol II and Pol III have five additional subunits forming a catalytic core that contains 10 subunits, as shown in Figure \(\PageIndex{9}\). The core has a characteristic crab-claw shape, which encloses a central cleft that harbors the DNA, and has two channels, one for the substrate NTPs and the other for the RNA product. Two ‘pinchers’, called the ‘clamp’ and ‘jaw’ stabilize the DNA at the downstream end and allow the opening and closing of the cleft. For transcription to occur, the enzyme has to maintain a transcription bubble with separated DNA strands, facilitate the addition of nucleotides, translocate along the template, stabilize the DNA:RNA hybrid, and finally allow the DNA strands to reanneal. This is achieved by a number of conserved elements in the active site, which include the fork loop(s), rudder, wall, trigger loop, and bridge helix.
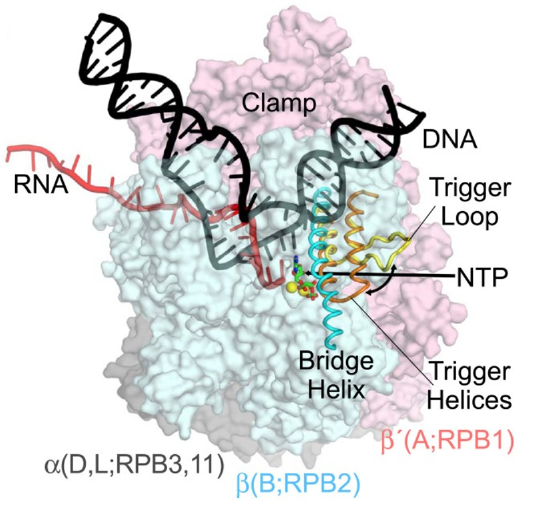
DNA (black) is melting into a transcription bubble that allows template-strand pairing with RNA (red) in a 9-10 base pair RNA-DNA hybrid. The bridge helix (cyan) and trigger loop/helices (yellow/orange) lie on the downstream side of the active site. The presumed path of the NTP entry is indicated by the straight arrow. Interconversion of the trigger loop and trigger helices is indicated by the curved arrow. The RNA polymerase subunits are shown as semi-transparent surfaces with the identities of orthologous subunits in bacteria (α, β, and β', gray, blue, and pink, respectively), archaea (D, L, B, and A), and eukaryotic RNA polymerase II (RPB3, 11, RPB2, RPB1) indicated. The active site Mg2+ ions are shown as yellow spheres, and α,β-methylene-ATP in green and red.
Table \(\PageIndex{1}\) shows RNA polymerase (RNA pol) subunit composition in bacteria, archaea, and, yeast and plants (both eukaryotes).
| Bacteria | Archaea | RNA pol I | RNA pol II | RNA pol III | RNA pol IV (plants) | RNA pol V (plants) |
|---|---|---|---|---|---|---|
| β | Rpo1 (RpoA) | RPA190 | RPB1 | RPC160 | NRPD1 | NRPE1 |
| β' | Rpo2 (RpoB) | RPBA135 | RPB2 | RPC128 | NRPD/E2 | NRPD/E2 |
| α | Rpo3 (RpoD) | RPAC40 | RPB3 | RPAC40 | RPB3 [1] | RPB3 [1] |
| α | Rpo11 (RpoL) | RPAC19 | RPB11 | RPAC19 | RPB11 | RPB11 |
| ω | Rpo6 (RpoK) | RPB6 | RPB6 | RPB6 | RPB6 [1] | RPB6 |
| Rpo5 (RpoH) | RPB5 | RPB5 | RPB5 | RPB5 [3] | NRPES5 | |
| Rpb8 (RpoG)* | RPB8 | RPB8 | RPB8 | RPB8 [1] | RPB8 [1] | |
| Rpo10 (RpoN) | RPB10 | RPB10 | RPB10 | RPB10 | RPB10 | |
| Rpo12 (RpoP) | RPB12 | RPB12 | RPB12 | RPB12 | RPB12 | |
| Rpo4 (RpoF) | RPA14 | RPB4 | RPC17 | NRPD/E4 | NRPD/E4 | |
| Rpo7(RpoE) | RPA43 | RPB7 | RPC25 | NRPD7 [1] | NRPE7 | |
| RPA12 | RPB9 | RPC11 | NRPD9b | RPB9 | ||
| Rpo13* | ||||||
| RPA49 | RPC53 | |||||
| RPA34.5 | RPC37 | |||||
| RPC82 | ||||||
| RPC34 | ||||||
| RPC31 |
Table \(\PageIndex{1}\): RNA polymerase (RNA pol) subunit composition. Abel, C., Verónica, M., I., G. A. , & Francisco, N. (2017). Subunits Common to RNA Polymerases. In (Ed.), The Yeast Role in Medical Applications. IntechOpen. https://doi.org/10.5772/intechopen.70936. Creative Commons Attribution 3.0 License
Schematic representations of the structure of the eukaryotic RNA pols I, II and III are shown in Figure \(\PageIndex{10}\).
Figure \(\PageIndex{10}\): Schematic representation of the structure of the RNA pols I, II and III. Each RNA pol common subunit is indicated in grey. The numbers correspond to each subunit are indicated in Subunits Common to RNA Polymerases. Abel et al, ibid.
RNA polymerases must bind to DNA, and to host of transcription factors (TF) necessary for specific and regulatable transcription. (Note: RNAP is not considered a transcription factor.) The comparative structures of RNAP I-III are shown in Figure \(\PageIndex{11}\). The "stalk" is a structural feature found in eukaryotes but not in prokaryotes. The figure focuses mostly on a comparison of RNAP I and RNAP III.
Figure \(\PageIndex{11}\): Comparison of RNAPI, II and III structures and transcription factors. Turowski TW and Boguta M (2021) Specific Features of RNA Polymerases I and III: Structure and Assembly. Front. Mol. Biosci. 8:680090. doi: 10.3389/fmolb.2021.680090. Creative Commons Attribution License (CC BY).
Panel (A) shows the general architecture of RNAPII, consisting of the catalytic core and stalk. RNAPII core consists of a DNA binding channel, catalytic center, and assembly platform. RNAPII binds multiple transcription factors (TFs). Some TFs are homologous to additional subunits of specialized RNAPs (i.e., TFIIF).
Panel (B) shows the subunit composition of eukaryotic RNAPs. Human nomenclature is shown for comparison. Please note that the C-terminal region of Rpa49 subunit harbors a “tandem winged helix” which is predicted in TFIIE and that human RNAPIII RPC7 subunit is coded by two isoforms α and β. The question mark indicates the name is unconfirmed.
Panel (C) shows the subunit composition of yeast RNAPI.
Panel (D) shows a Model of the RNAPI pre-initiation complex, showing an early intermediate with visible Rrn3 and core factor (CF). TATA-binding protein (TBP) and upstream-associated factor (UAF) are added schematically.
Panel (E) shows the subunit composition of yeast RNAPIII.
Panle (F) shows atomic model of RNAPIII pre-initiation complex with TFIIIB. The Rpc82/34/31 heterotrimer is involved in initiation and marked in green as in E. TFIIIC is added schematically. PDB: 5C4X, 5FJ8, 4C3J, 6EU0, and 6TPS
The stalks have two proteins that are not as homologous as the core subunits. These are highlighted in blue in Panel B of Figure \(\PageIndex{11}\). RNAP I and III have additional subunits compared to RNAP II. Overall RNAP I-III have 14, 12, and 17 subunits, respectively. RNAP I and III appear to have integrated transcription-like factors into their core enzyme. In contrast, RNAP II (which transcribes DNA to form messenger RNA), binds to discrete and separate transcription factors to form a preinitiation complex (PIC)which we will discuss below.
Transcription Factors and the Preinitiation Complex (PIC)
Unlike prokaryotic systems which can initiate the recruitment of RNAP holoenzymes directly onto the DNA promoter regions and mediate the conversion of RNAP to the open conformation, eukaryotic RNA polymerases require a host of additional general transcription factors (GTFs), to enable this process. Here we will focus on the activation of RNA Polymerase II as an example of the complexity of eukaryotic transcription initiation.
Class II gene transcription in eukaryotes is a tightly regulated, essential process controlled by a highly complex multicomponent machinery. A plethora of proteins, more than a hundred in humans, are organized in very large multiprotein assemblies that include a core of General Transcription Factors (GTFs). The GTFs include the factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, RNA polymerase (RNA pol II), as well as a large number of diverse complexes that act as co-activators, co-repressors, chromatin modifiers, and remodelers, as shown in Figure \(\PageIndex{12}\). Class II gene transcription is regulated at various levels: while assembling on chromatin, before and during transcription initiation, throughout elongation and mRNA processing, and termination. A host of activators and repressors has been reported to regulate transcription, including a central multisubunit complex called the Mediator that helps in the recruitment of GTFs and the activation of RNA Pol II. Here we will focus on the formation of the GTFs that make up the core preinitiation complex (PIC) during transcriptional activation.
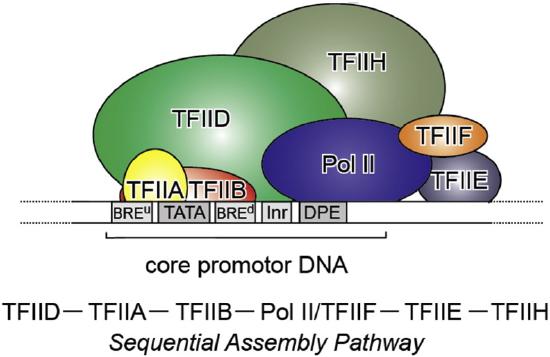
Class II gene transcription in humans is brought about by over a hundred polypeptides assembling on the core promoter of protein-encoding genes, which then give rise to mRNA. A PIC on a core promoter is shown in a schematic representation. PIC contains, in addition to promoter DNA, the GTFs (TFIIA, B, D, E, F, and H), and RNA Pol II. PIC assembly is thought to occur in a highly regulated, stepwise fashion, as indicated. TFIID is among the first GTFs to bind the core promoter via its TATA-box Binding Protein (TBP) subunit. Nucleosomes at transcription start sites contribute to PIC assembly, mediated by signaling through epigenetic marks on histone tails. The Mediator (not shown) is a further central multiprotein complex identified as a global transcriptional regulator. TATA = TATA-box DNA; BREu = B recognition element upstream; BREd = B recognition element downstream; Inr = Initiator; DPE = Down-stream promoter element. Figure from:
Transcription of RNA pol II-dependent genes is triggered by the regulated assembly of the Preinitiation Complex (PIC). PIC formation starts with the binding of TFIID to the core promoter. TFIID is a large megadalton-sized multiprotein complex with around 20 subunits made up of 14 different polypeptides: the TATA-box binding protein (TBP) and the TBP-associated factors (TAFs) (numbered 1–13), as shown in Figure \(\PageIndex{13}\). Some of the TAF subunits are present in two copies. A key feature in TAFs is the histone fold domain (HFD), which is present in 9 out of 13 TAFs in TFIID. The HFD is a strong protein–protein interaction motif that mediates specific dimerization. The HFD-containing TAFs are organized in discrete heterodimers, with the exception of TAF10, which is capable of forming dimers with two different TFIID components, TAF3 and TAF8. HFDs and several other structural features of TBP and the TAFs are well conserved between species.
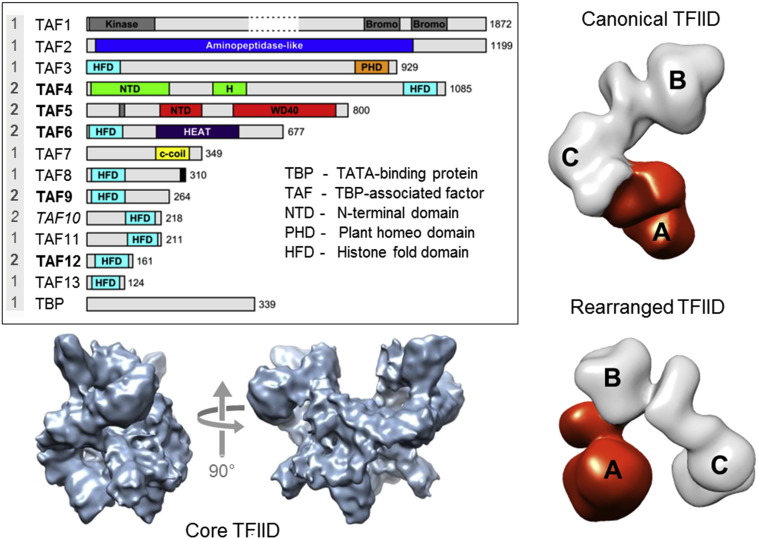
TFIID is a large megadalton-sized multiprotein complex comprising about 20 subunits made up of 14 different polypeptides. The constituent proteins of TFIID, TBP and the TAFs, are shown in a schematic representation depicted as bars (inset, left). Structured domains are marked and annotated. The presumed stoichiometry of TAFs and TBP in the TFIID holo-complex is given (far left, gray underlaid). TAF10 (in italics) makes histone fold pair separately with both TAF3 and TAF8. TAFs present in a physiological TFIID core complex extracted from eukaryotic nuclei are labeled in bold. The architecture of the TFIID core complex (EMD-2230) determined by cryo-EM is shown (bottom left) in two views related by a 90° rotation (arrows). The holo–TFIID complex is characterized by remarkable structural plasticity. Two conformations, based on cryo-EM data (EMD-2284 and EMD-2287), are shown on the right, a canonical form (top) and a more recently observed rearranged form (bottom). In the rearranged conformation, lobe A (colored in red) migrates from one extreme end of the TFIID complex (attached to lobe C) all the way to the other extremity (attached to lobe B).
TFIID was shown to adopt an asymmetric, horse-shoe shape with three almost equal-sized lobes (A, B, and C), exhibiting a considerable degree of conformational flexibility with at least two distinct conformations (open and closed), as shown in Figure \(\PageIndex{12}\). The TBP component of TFIID binds with a specific DNA sequence called the TATA box. This DNA sequence is found around 30 base pairs upstream of the transcription start site in many eukaryotic gene promoters. When TBP binds to a TATA box within the DNA, it distorts the DNA by inserting amino acid side chains between base pairs, partially unwinding the helix, and doubly kinking it. The distortion is accomplished through a great amount of surface contact between the protein and DNA. TBP binds with the negatively charged phosphates in the DNA backbone through positively charged lysine and arginine amino acid residues. The sharp bend in the DNA is produced through the projection of four bulky phenylalanine residues into the minor groove. As the DNA bends, its contact with TBP increases, thus enhancing the DNA-protein interaction. The strain imposed on the DNA through this interaction initiates the melting, or separation, of the strands. Because this region of DNA is rich in adenine and thymine residues, which base-pair through only two hydrogen bonds, the DNA strands are more easily separated.
The role of TAFs is complicated. Take for example TAF11 and TAF13. These act as competitive inhibitors of TBP to the TATA (Pribnow) box as well as TAF1 which somewhat mimics the structural features of the Pribnow box. TAF11/TAF13 binds to the DNA surface where TBP binds. suggesting a novel regulation of TFIID. These interactions are illustrated in Figure \(\PageIndex{14}\).
Figure \(\PageIndex{14}\): Novel TFIID regulatory state comprising TAF11/TAF13/TBP. Kapil Gupta,et al. (2017) Architecture of TAF11/TAF13/TBP complex suggests novel regulation properties of general transcription factor TFIID eLife 6:e30395. https://doi.org/10.7554/eLife.30395. Creative Commons Attribution License
Given its role in transcribing the DNA for thousands of messenger RNAs, let's focus on the preinitation complex for RNAP II. Structures are known for the closed and open promoters. A key component is TFH (see Figure \(\PageIndex{12}\)). TFIIH opens the DNA for transcription. As if we need to complicate the structure of the preinitiation complex even more, it turns out the TFIIH is not a single dark green cartoon as shown in Figure 12, but a rather large complex itself. Its structure is shown in Figure \(\PageIndex{15}\).
Figure \(\PageIndex{15}\): Structure of the TFIIH core complex. Greber et al. (2019). The complete structure of the human TFIIH core complex. eLife 8:e44771. https://doi.org/10.7554/eLife.44771. Creative Commons Attribution License
Panes (A, B, C) shows three views of the structure of the TFIIH core complex and MAT1. Subunits are color-coded and labeled (in color); individual domains are labeled (in black) and circled if needed for clarity.
Panel (D) shows the domain-level protein-protein interaction network between the components of the TFIIH core complex and MAT1 derived from the interactions observed in our structure. Proteins are shown with the same colors as in A and major unmodeled regions are shown in grey. Abbreviations: CTD: C-terminal domain; DRD: DNA damage recognition domain; FeS: iron sulfur cluster domain; NTD: N-terminal domain; vWFA: von Willebrand Factor A.
The largest subunits are DNA-dependent helicases/translocases/ATPases XPB and XPD, which are bridged by MAT1. It appears that XPB starts and propagates a twist in the DNA which propagates ot open the DNA 30 BP downstream of the TATA box. This mostly likely is followed by the dissociation of TFIIH and a stoppage in DNA twisting, which allows RNA transcription to start.
Figure \(\PageIndex{16}\) shows an interactive iCn3D model of the human/mouse/mastadenovirus C RNA polymerase II core pre-initiation complex with open promoter DNA (7NVU).
The following color schemes are used:
- MAT1 - orange
- TF2A - yellow
- TF2B - green
- TF2E - magenta
- TF2F- purple
- ATP-dependent translocase (helicase) subunit XPD - dark slate gray
- TBP (TATA box binding protein) - red
- TF2H - Pink
- 2H-XPB helicase - maroon
- NT-DNA - cyan
- template (T)-DNA - blue (Note: part of it is shown in a yellow cartoon in the interactive model)
- the SF4 cofactor is shown in spacefill with CPK colors
In summary, the binding of TFIID to the core promoter is followed by the recruitment of further GTFs and RNA pol II. Several lines of evidence suggest that this process occurs in a defined, stepwise order and undergoes significant restructuring. First, PIC adopts an inactive state, the “closed” complex, which is incompetent to initiate transcription. In addition to TFIID, TFIIH is also critical for the shift of RNA Pol II from the closed to the open conformation. TFIIH has an ATP-dependent translocase activity within one of its subunits, that opens up about 11 to 15 base pairs around the transcription start site by moving along one DNA strand inducing torsional strain, leading to conformational rearrangements and the positioning of single-stranded DNA to the active site of RNA pol II. In this “open” complex, RNA pol II can enter elongation to transcribe throughout a gene in a highly processive manner without dissociating from the DNA template or losing the nascent RNA.
In most eukaryotes, after synthesizing about 20–100 bases, RNA pol II can pause (Promoter proximal pause) and then disconnect from promoter elements and other components of the transcription machinery, giving rise to a fully functional elongation complex in a process called promoter escape. The promoter-bound components of the PIC, in contrast, remain in place, and thus only TFIIB, TFIIF, and RNA pol II need to be recruited for re-initiation, significantly increasing the transcription rate in subsequent rounds of transcription. Promoter escape is preceded by an abortive transcription in many systems, where multiple short RNA products of 3 to 10 bases in length are synthesized.
In addition to promoter elements within the DNA, enhancer elements are also important for the initiation of transcription. Promoters are defined as DNA elements that recruit transcription complexes for the synthesis of coding and non-coding RNA. Enhancers are defined as DNA elements that positively regulate transcription at promoters over long distances in a position- and orientation-independent manner. However, studies have revealed that many enhancers can recruit Pol II and initiate transcription of enhancer RNA (eRNA), thus blurring the functional distinction between enhancers and promoters (Figure 10.13).
Enhancer transcription produces relatively short ncRNA. Furthermore, transcription at enhancers is unstable and often leads to the termination of elongation. In contrast, transcription initiation at most Pol II promoters is stable and produces long mRNAs. Topological studies revealed that enhancers come in close proximity to target gene promoters during transcription activation. According to current gene activation models, the Mediator complex forms a physical bridge between distant regulatory regions and promoters, thereby promoting looping. Transcription of at least a subset of genes regulated by enhancers occurs in bursts indicating a discontinuous process of transcription complex recruitment, assembly, and/or conversion to elongation-competent forms. The bursting phenomenon suggests that enhancer/promoter contacts may be transient and infrequent, as shown in Figure \(\PageIndex{17}\).
Depicted are the steps involved in the recruitment of Pol II to SEs, assembly into elongation-competent transcription complexes, transcription initiation, and elongation, abortion and termination, and transfer to target genes. Transcription factors recruit Mediator and other co-regulators to SEs. Mediator recruits Pol II and assembles a fraction into elongation competent transcription complexes. Transcription is initiated by phosphorylation of the CTD. Early abortion and transcription termination conferred by Integrator releases Pol II, which is dephosphorylated and transferred to target gene promoters. Super Enhancer Element (SE).
Transcriptional Elongation and Termination
Prokaryotic Transcriptional Elongation
The rate of transcription elongation by E. coli RNAP is not uniform. RNA synthesis is characterized by pauses, some of which may be brief and resolved spontaneously, whereas others may lead to the transcription elongation complex (TEC) backtracking.
Elongation rate and pausing are determined by template sequence and RNA structure (e.g., stem-loops) and involve at least two components of the RNAP catalytic center, the bridge helix (BH) and trigger loop (TL). Elongation is proposed to occur in three steps, as shown in Figure \(\PageIndex{18}\). First, the TL folds in response to NTP binding. Mutational analyses indicate that this conformational change in the TL can be rate-limiting, and reflects the ability of the incoming NTP to bind to TEC. The second step is the incorporation of the NTP and the release of pyrophosphate. The third step involves the translocation of the RNAP down the DNA Template such that the next RNA nucleotide can be added to the nascent transcript.
The trigger loop hinges, bridge helix hinges, and bridge helix bending models are based on molecular dynamics simulations. At the top of the figure, diagrams of the closed TEC, the closed product TEC (after chemistry), and the translocating TEC are shown. DNA is grey; RNA is red; the NTP substrate (or incorporated NMP and pyrophosphate) is blue; the trigger loop (TL) is purple; the bridge helix (BH) is yellow. Interpretations of simulations are shown schematically below. Simulations indicate trigger loop hinges H1 and H2, bridge helix hinges H3 and H4 and bridge helix bend modes B1 (straighter) and B2 (more sharply bent).
Backtracking of TEC may take place after a brief pause in transcription, caused by the thermodynamic properties of nucleic acids sequences surrounding the elongation complex. In addition, misincorporation events render elongation complexes prone to backtracking by at least one bp. In this case, the rescue from backtracking through the cleavage of the 3' end of the erroneous transcript also may be seen as a proofreading reaction. Any backtracking event causes a pause or arrest of transcription elongation, which may limit its overall rate (the average speed of RNAP along the template) or the processivity (the fraction of RNAP molecules reaching the end of the gene).
While the general structure of the elongation complex (the transcription bubble, the RNA-DNA hybrid) remains unchanged during backtracking, the extension of RNA becomes impossible in this conformation. However, such complexes can be resolved by the hydrolytic activity of RNAP, which cleaves the phosphodiester bond in the active center of the backtracked complex, producing a new RNA 3' end in the active center. For single base backups, the hydrolytic reaction is catalyzed by a flexible domain of RNAP located in the secondary channel called the Trigger Loop (TL) and the two metal ions of the active center.
Longer sequences of backtracked TEC can restart when acted upon by GreA/B factors, which restore the 3'-end of the nascent transcript to the active center. GreA and GreB are transcript cleavage factors that act on backtracked elongation complexes. When Gre factors are bound in the secondary channel, Gre factors displace the TL from the active center, as shown in Figure \(\PageIndex{19}\). The displacement switches off the relatively slow TL-dependent intrinsic transcript hydrolysis, and imposes the highly efficient Gre-assisted hydrolysis. This efficiency is thought to be due to the stabilization of the second catalytic Mg2+ ion and an attacking water molecule by the Gre factors.
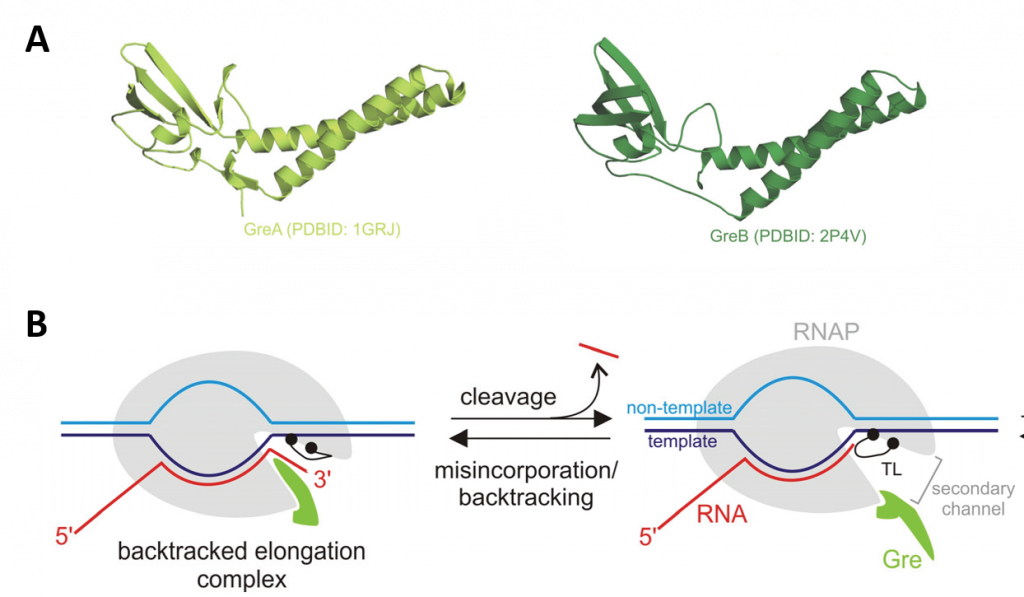
Panel (A) shows a ribbon diagram of the GreA and GreB proteins.
Panel (B) shows the mode of functioning of Gre factors. The Gre factor is bound to the active elongation complex but does not impose hydrolytic activity on it. Upon backtracking or misincorporation, the Gre factor protrudes its coiled-coil domain through the secondary channel of RNAP (shown in the lefthand diagram), where it substitutes for the catalytic domain Trigger Loop (TL). This substitution switches off the slow TL-dependent phosphodiester bond hydrolysis and, and instead, facilitates highly efficient Gre-dependent hydrolysis. After the resolution of the backtracked complex through RNA cleavage, the elongation complex returns to the active conformation, and the Gre factor gives way to the TL, which can now continue the catalysis of RNA synthesis (shown in the right-hand diagram). The controlled switching between Gre and the TL eliminates possible interference of Gre with the RNA synthesis.
Prokaryotic Transcriptional Termination
Transcription termination determines the ends of transcriptional units by disassembling the transcription elongation complex (TEC), thereby releasing RNA polymerases and nascent transcripts from DNA templates. Failure in termination causes transcription readthrough, which yields wasteful and possibly harmful intergenic transcripts. It can also perturb the expression of downstream genes when the unterminated TEC sweeps transcription initiation complexes off their promoters or collides with RNA polymerases that transcribe opposite strands.
Transcriptional termination in prokaryotes can be template-encoded and factor-independent (intrinsic termination), or require accessory factors, such as Rho, Mfd, and DksA. Intrinsic termination occurs at specific template sequences - an inverted repeat followed by a run of A residues. Termination is driven by the formation of a short stem-loop structure in the nascent RNA chain, as shown in Figure \(\PageIndex{20}\). RNA synthesis arrests and TEC dissociates at the 7th and 8th U of the run. Formation of the stem-loop dissociates the weak rU:dA hybrid. Stem-loop formation is hindered by upstream complementary RNA sequences that compete with the downstream portion of the stem, as well as by RNA: protein interactions in the RNA exit channel. Intrinsic termination depends critically upon timing. Hairpin folding and transcription of the termination point must be coordinated, so that the complete hairpin is formed by the time RNAP transcribes the termination point. The size of the stem, the sequence of the stem, and the length of the loop all affect termination efficiency.
The bridge α-helix in the β' subunit borders the active site and may have roles in both catalysis and translocation. Mutations in the YFI motif (β' 772-YFI-774) affect intrinsic termination as well as pausing, fidelity, and translocation of RNAP. One mutation, F773V, abolishes the activity of the λ tR2 intrinsic terminator, although neighboring mutations have little effect on termination. Modeling suggests that this unique phenotype reflects the ability of F773 to interact with the fork domain in the β subunit.
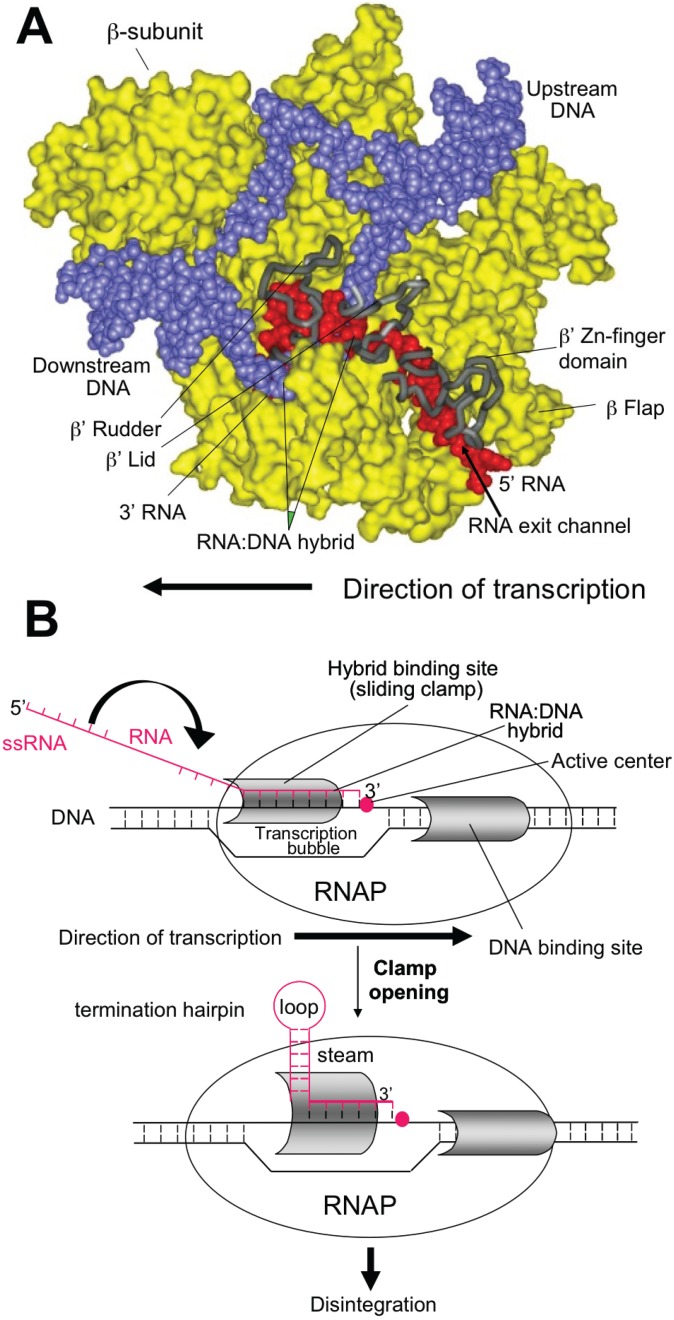
Panel (A) shows the open conformation of the RNAP during transcriptional elongation. RNAP is shown in yellow, the DNA template in blue, and the nascent RNA in red. Key elements of the RNAP RNA exit channel are shown in grey and labeled as indicated.
Panel (B) shows the extension of the nascent RNA through the RNAP exit channel and the potential for forming the RNA hairpin structure when enough length has been achieved.
Panel (C) shows the clamp opening and disintegration of the TEC when the RNA hairpin structure is encountered at the transcriptional bubble.
Figure \(\PageIndex{21}\) shows an interactive iCn3D model of the T. thermophilus RNAP polymerase elongation complex with the NTP substrate analog (2O5J). (long load time)
Transcriptional termination can also be dependent upon accessory factors, such as the Rho protein. Transcription termination factor Rho is an essential protein in E. coli first identified for its role in transcription termination at Rho-dependent terminators, and is estimated to terminate ~20% of E. coli transcripts. The rho gene is highly conserved and nearly ubiquitous in bacteria. Rho is an RNA-dependent ATPase with RNA:DNA helicase activity, and consists of a hexamer of six identical monomers arranged in an open circle, as shown in Figure \(\PageIndex{22}\).
Rho binds to single-stranded RNA in a complex multi-step pathway that involves two distinct sites on the hexamer. The primary binding site (PBS), distributed on the N-terminal domains around the hexamer (cyan), ensures initial anchoring of Rho to the transcript at a Rut (Rho utilization) site, a∼70 nucleotides (nt) long, cytidine-rich and poorly-structured RNA sequence. Each Rho monomer contains a subsite capable of binding specifically the base residues of a 5′-YC dimer (Y being a pyrimidine). Biochemical and structural data suggest that Rho initially binds to RNA in an open, ‘lock-washer’ conformation that closes into a planar ring as RNA transfers to the central cavity. There, the ssRNA contacts an asymmetric secondary binding site (SBS) (green), and this step, which presumably is rate-limiting for the overall reaction, leads to motor activation. Upon hydrolysis of ATP, the ssRNA is pulled upon conformational changes of the conserved Q and R loops of the SBS, leading to Rho translocation, and ultimately promoting RNA polymerase (RNAP) dissociation. The molecular mechanism of Rho translocation based on single-molecule fluorescence methods appears to be tethered tracking. The tethered tracking model postulates that Rho maintains its contacts between the PBS and the loading (Rut) site upon translocation (Panel B). This mechanism would allow Rho to maintain its high-affinity interaction with Rut, and implies the growth of an RNA loop between the PBS and the SBS upon translocation.
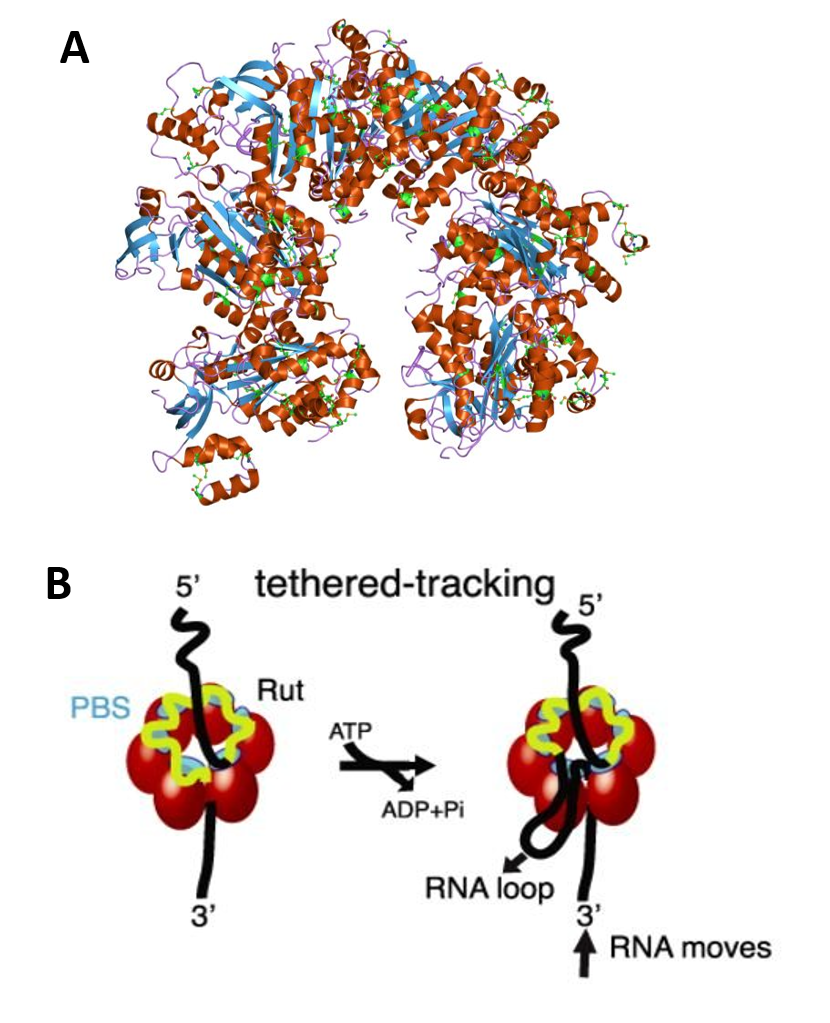
Panel (A) shows the molecular structure of the Rho protein (PDB 1pv4)
Panel (B)shows how Rho assembles as a homo-hexameric ring (red spheres or tetragons), with RNA (black/yellow curve) binding to the primary binding sites (PBS, cyan) and the secondary binding sites inside the ring (SBS, green), where ATP-coupled translocation takes place. The Rut-specific binding site is depicted in yellow. The tethered-tracking model proposed that Rho translocates RNA while maintaining interactions between PBS and Rut. This model requires the formation of a loop that would shorten the extension of RNA upon translocation. Figure modified from:
Figure \(\PageIndex{23}\) shows an interactive iCn3D model of the E. Coli Rho transcription termination factor in complex with ssRNA substrate and ANPPNP (1PVO).
The six subunits of the hexamer are shown in alternating slate gray and light gray. The di-ribonucleotides (5'-R(P*UP*C)-3') are shown in spacefill and colored CPK. ANPPNP is shown in spacefill yellow.
Figure \(\PageIndex{24}\) shows an interactive iCn3D model of the closed ring structure of the E. Coli Rho transcription termination factor in a complex with nucleic acid in the motor domains (2HT1).
The six subunits of the hexamer are again shown in alternating slate gray and light gray. The ssRNAs are shown in spacefill with the backbones in one color and the bases in CPK colors.
Figure \(\PageIndex{25}\) shows an interactive iCn3D model of the E. coli Rho-dependent Transcription Pre-termination Complex (6XAS) (long load).
- The RNA polymerase subunits (α2ββω) are shown in light cyan
- The six subunits of the Rho hexamer are again shown in alternating slate gray and light gray
- NusA is shown in magenta
- Both DNA strands are shown in spacefill with the backbone blue and the bases red
- Only part of the RNA is shown (spacefill, backbone yellow, bases CPK), so it appears discontinuous
Eukaryotic Transcriptional Termination
In eukaryotes, termination of protein-coding gene transcription by RNA polymerase II (Pol II) usually requires a functional polyadenylation (pA) signal, typically a variation of the AAUAAA hexamer. Nascent pre-mRNA is cleaved and the 5′ fragment is polyadenylated at the pA site shortly downstream from the hexamer by cleavage and pA factors (CPFs). Two mechanisms have been suggested for pA-dependent transcription termination. In the allosteric model, the pA signal and/or other termination signals bind with the pA signal downstream region (PDR) and induce reorganization of the Pol II complex. This includes the association or dissociation of endonuclease components such as the CPFs. This causes conformational changes in Pol II and TEC disassembly ensues. In the kinetic model, also known as the “torpedo” model, cleavage at the pA site separates the pre-mRNA from the TEC, which continues synthesizing a downstream nascent transcript. This new transcript is a substrate of XRN2/Rat1p, a processive 5′-to-3′ exoribonuclease that catches up with, and disassembles, the TEC by an unknown mechanism.
The two pA-dependent models are not mutually exclusive, and unified models have been proposed. Loosely conserved pA signal sequences downstream of protein-coding genes bind to components of the polyadenylation factor (CF1) complex leading to the assembly of the cleavage and polyadenylation machinery. Termination is coupled to cleavage in a manner that has not yet been completely resolved, however, one of the major factors involved in yeast pA termination is the endonuclease, Ysh1. For example, the depletion of Ysh1 blocks TEC dissociation, but does not cause substantial readthrough at the termination site (Fig. 26.1.18 A&B). These results suggest that Ysh1 does not directly cause the pausing that occurs in the allosteric termination pathway, but rather plays a role in the dissociation of the Pol II complex from the DNA template, as shown in Figure \(\PageIndex{26}\). It should be noted that not all pA-dependent termination is dependent on Ysh1 and that other mechanisms of pA-mediated termination still remain to be elucidated.
Panel (A) shows the elongating Pol II (green) terminates pA transcripts (A) after an allosteric change (red) that reduces processivity.
Panel (B) shows the depletion of Ysh1 leads to minimally extended readthrough transcripts but does not block the allosteric change in Pol II.
Panel (C) shows how Nrd1 and Nab3 binding recruits Sen1 for termination of non-pA transcripts.
Panel (D) shows Pol II elongation complex lacking Nrd1 does not recognize termination sequences in the nascent transcript and thus does not facilitate the allosteric transition in Pol II. This leads to a processive readthrough.
Panel(E) shows how Nrd1 and Nab3 recognize terminator sequences allowing the allosteric change in Pol II but depletion of Sen1 blocks the removal of Pol II from the template. Figure from:
The mechanisms of termination of Pol II-mediated transcription differ for coding and non-coding transcripts. Coding transcripts and possibly some stable uncharacterized transcripts (SUTs) are nearly always processed at the 3′-end by the cleavage and polyadenylation (pA) machinery and are processed by the pA-dependent termination mechanisms described above. In contrast, ncRNAs are terminated and processed by an alternative pathway that, in yeast, requires the RNA-binding proteins Nrd1 and Nab3, as well as, the RNA helicase Sen1. Nrd1 and Nab3 recognize RNA sequence elements downstream of snoRNAs and CUTs and this leads to the association of a complex that contains the DNA/RNA helicase Sen1 and the nuclear exosome. The nuclear exosome is a complex of ribonucleases with 3' to 5' exonuclease and endonuclease activity. It functions to degrade unstable or incorrect RNA transcripts.
Both Nrd1 and Sen1 depletion lead to readthrough transcription of ncRNAs, suggesting their importance in non-pA-dependent transcription termination (Fig 26.1.18 C & D). Furthermore, depletion of Nrd1 also causes the accumulation of longer readthrough ncRNAs, suggesting its role in trafficking ncRNAs to the nuclear exosome following termination.
- Parker, N., Schneegurt, M., Thi Tu, A-H., Lister, P., Forster, B.M. (2019) Microbiology. Openstax. Available at: https://opentextbc.ca/microbiologyopenstax/
- Palazzo, A., and Lee, E.S. (2015) Non-coding RNA: what is function and what is junk? Frontiers in Genetics 6:2 Available at: file:///C:/Users/flatt/AppData/Local/Temp/fgene-06-00002.pdf
- Wikipedia contributors. (2020, July 9). RNA. In Wikipedia, The Free Encyclopedia. Retrieved 15:30, August 6, 2020, from https://en.Wikipedia.org/w/index.php?title=RNA&oldid=966784317
- Burenina, O.Y., Oretskaya, T.S., and Kubareva, E.A. (2017) Non-Coding RNAs As Transcriptional Regulators in Eukaryotes. Acta Naturae 9(4):13-25. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5762824/
- Khatter, H., Vorlander, M.K., and Muller C.W. (2017) RNA polymerase I and III: similar yet unique. Current Opinion in Structural Biology 47:88-94. Available at: https://www.sciencedirect.com/science/article/pii/S0959440X17300313
- Wikipedia contributors. (2020, May 8). Sigma factor. In Wikipedia, The Free Encyclopedia. Retrieved 17:50, August 7, 2020, from https://en.Wikipedia.org/w/index.php?title=Sigma_factor&oldid=955570499
- Bae, B., Felkistov, A., Lass-Napiokowska, A., Landick, R., and Darst, S.A. (2015) Structure of a bacterial RNA polymerase holoenzyme open protomer complex. eLife 4:e08504. Available at: https://elifesciences.org/articles/08504
- Petrenko, N., Jin, Y., Dong, L., Wong, K.H., and Struhl, K. (2019) Requirements for RNA polymerase II preinitiation complex formation in vivo. eLife 8:e43654. Available at: https://elifesciences.org/articles/43654
- Gupta, K., Sari-Ak, D., Haffke, M., Trowitzsch, S., and Berger, I. (2016) Zooming in on transcription preinitiation. J Mol Biol. 428(12):2581-2591. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4906157/
- Wikipedia contributors. (2020, April 17). TATA-binding protein. In Wikipedia, The Free Encyclopedia. Retrieved 14:54, August 8, 2020, from https://en.Wikipedia.org/w/index.php?title=TATA-binding_protein&oldid=951583592
- Patel, A.B., Greber, B.J., and Nogales, E. (2020) Recent insights into the structure of TFIID, its assembly, and its binding to core promoter. Curr Op Struct Bio 61:17-24. Available at: https://www.sciencedirect.com/science/article/pii/S0959440X19301113#fig0010
- Ruff, E.F., Record, Jr., M.T., Artsimovitch, I., (2015) Initial events in bacterial transcription initiation. Biomolecules 5(2):1035-1062. Available at: https://www.mdpi.com/2218-273X/5/2/1035/htm
- Kireeva, M., Opron, K., Seibold, S., Domecq, C., Cukier, R.I., Coulombe, B., Kashlev, M., and Burton, Z. (2102) Molecular dynamics and mutational analysis of the catalytic and translocation cycle of RNA polymerase. BMC Biophysics 5(1):11. Available at: https://www.researchgate.net/publication/225281979_Molecular_dynamics_and_mutational_analysis_of_the_catalytic_and_translocation_cycle_of_RNA_polymerase/figures?lo=1
- Washburn, R.S., and Gottesman, M.E. (2015) Regulation of transcription elongation and termination. Biomolecules 5(2):1063-1078. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4496710/pdf/biomolecules-05-01063.pdf
- Zenkin, N., and Yuzenkova, Y. (2015) New insights into the functions of transcription factors that bind the RNA polymerase secondary channel. Biomolecules 5(3):1195-1209. Available at: https://www.mdpi.com/2218-273X/5/3/1195/htm
- Gocheva, V., LeGall, A., Boudvillain, M., Margeat, E., and Nollmann, M. (2015) Direct observation of the translocation mechanism of transcription termination factor Rho. Nuc Acids Res 43(1):10.1093. Available at: https://www.researchgate.net/publication/272162172_Direct_observation_of_the_translocation_mechanism_of_transcription_termination_factor_Rho
- Miki, T.S., Carl, S.H., and Groβhans, H. (2017) Two disctinct transcription termination modes dictated by promoters. Genes & Dev 31:1-10. Available at: https://www.researchgate.net/publication/320350041_Two_distinct_transcription_termination_modes_dictated_by_promoters
- Gurumurthy, A., Shen, Y., Gunn, E.M., Bungert, J. (2018) Phase separation and transcription regulation: Are Super-Enhancers and Locus Control Regions primary sites of transcription complex assembly? BioEssays 1800164. Available at: https://www.researchgate.net/publication/329331157_Phase_Separation_and_Transcription_Regulation_Are_Super-Enhancers_and_Locus_Control_Regions_Primary_Sites_of_Transcription_Complex_Assembly
- Suñé-Pou, M., Prieto-Sánchez, Boyero-Corral, S., Moreno-Castro, C., El Yousfi, Y., Suñé-Negre, J.M., Hernández-Munain, C., and Suñé, C. (2017) Targeting splicing in the treatment of human disease. Genes 8(3):87. Available at: https://www.mdpi.com/2073-4425/8/3/87/htm
- Schaughency, P., Merran, J., and Corden J.L. (2014) Genome-wide mapping of yeast RNA polymerase II termination. PLOS Genetics 10(10):e1004632 Available at: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1004632
- Nourse, J., Spada, S., and Danckwardt, S. (2020) Emerging roles of RNA 3'-end cleavage and polyadenylation in pathogenesis, diagnosis, and therapy of human disorders. Biomolecules 10(6):915. Available at: https://www.mdpi.com/2218-273X/10/6/915/htm
- Wikipedia contributors. (2020, July 30). Five-prime cap. In Wikipedia, The Free Encyclopedia. Retrieved 05:53, August 11, 2020, from https://en.Wikipedia.org/w/index.php?title=Five-prime_cap&oldid=970240533
- Cortes, T. and Cox, R.A. (2015) Transcription and translation of the rpsJ, rplN and rRNA operons of the tubercle bacillus. Microbiology (2015) 161:719-728. Available at: https://www.microbiologyresearch.org/docserver/fulltext/micro/161/4/719_mic000037.pdf?expires=1597159574&id=id&accname=guest&checksum=6FFC9C066EF41C7799FAE843CE94C49F
- Hein, P.P. and Landick, R. (2010) The bridge helix coordinates movements of modules in RNA polymerase. BMC Biology 8:141. Available at: https://bmcbiol.biomedcentral.com/articles/10.1186/1741-7007-8-141
- Gonatopoulos-Pournatzis, T., and Cowling, V.H. (2014) Cap-binding complex (CBC). Biochem. J. 457:231-242. Available at: https://www.researchgate.net/publication/259392894_Cap-binding_complex_CBC




Opt.png?revision=1&size=bestfit&width=320&height=349)
.png?revision=1&size=bestfit&width=597&height=401)
.png?revision=1&size=bestfit&width=430&height=369)
.png?revision=1&size=bestfit&width=483&height=474)
.png?revision=1&size=bestfit&width=388&height=370)
.png?revision=1&size=bestfit&width=495&height=341)