20.6: Biosynthesis of Starch, Sucrose and Cellulose
- Page ID
- 41604
Now that we have seen how carbon is captured and fixed into 3C trioses, which can be converted to fructose and glucose and their derivative, we can now explore the synthesis of the key plant carbohydrates we all know, sucrose, starch, and cellulose.
The source for the organization and some of the text derives from Sindayigaya and Longhini. https://www.peoi.org/Courses/Courses...chem/biochem18. Creative Commons - https://creativecommons.org/licenses...sa/3.0/deed.en
Sucrose Synthesis
Sucrose is a disaccharide of glucose and fructose with an acetal link between the anomeric carbons to form the nonreducing sugar O-α-D-glucopyranosyl-(1→2)-β-D-fructofuranoside. Its structure is shown in Figure \(\PageIndex{1}\).
Figure \(\PageIndex{1}\): Sucrose, a disaccharide of glucose and fructose
Sucrose can be considered a transport form of carbon, much like ketone bodies are transport forms of fatty acids. As noted above, the link between the sugars is between the anomeric C-1 link of glucose and the anomeric C-2 link of fructose. As such, it is not cleaved by typical carbohydrate-cleaving enzymes like amylases. Also, it doesn't react with proteins like other sugars with free cyclic hemiacetals that can open and form reactive aldehydes. For example, the cyclic monosaccharide glucose, with its anomeric carbon in a readily reversible hemiacetal link, can form covalent bonds to amine groups in proteins such as hemoglobin, which forms the glycosylated version of HbA1c, as shown in Figure \(\PageIndex{2}\).
Figure \(\PageIndex{2}\): Reactions of cyclic hemiacetal sugars with amines. https://en.wikipedia.org/wiki/Glycated_hemoglobin. Creative Commons Attribution-ShareAlike License 3.0
The reaction proceeds through the formation of a Schiff base followed by a rearrangement. HbA1c is a marker of diabetes. These features of sucrose may explain its choice as a key form of synthesized carbohydrate in plants.
Sucrose is synthesized in the cytosol, as shown in Figure \(\PageIndex{3}\).
Figure \(\PageIndex{3}\): Synthesis of sucrose in plants.
We have seen the enzymes that catalyze these reactions through G1P before in the glycolytic and gluconeogenesis pathways. Glucose-1-phosphate is then converted to UDP-glucose, which reacts with fructose-6-phosphate to form sucrose 6-phosphate and UDP, catalyzed by the enzyme sucrose 6-phosphate synthase. The last reaction is the hydrolysis of sucrose 6-phosphate by sucrose 6-phosphatase, which allows the export and transport of sucrose.
In the reaction scheme above, G1P is converted to UDP-Glc through the following reaction:
G1P + H+ + UTP ↔ UDP-Glc + Pi
This reaction is just mildly favored thermodynamically.
Before we study the structure of the enzyme, we will first discuss the regulation of the sucrose synthesis pathway and discuss the generic mechanism of glycosyltransferases.
Regulation
Carbon capture in the light reaction of photosynthesis leads to sucrose (for transport) and starch synthesis. Which product(s) result depends on key regulatory steps. Remember of the 6 trioses formed in the Calvin cycle, 5 are returned into the cycle for the synthesis of ribulose 1,5-bisphosphate, and only 1 is used for the synthesis of sucrose and starch. If too much is removed, the cycle slows. If not enough is used for starch and sucrose synthesis, Pi, which is moved into the stroma from the cytoplasm by an important translocator (see section 20.3) would run low.
The key cytosolic regulatory step is catalyzed by fructose 1,6-bisphosphatase (FBPase-1) and also a unique plant enzyme, PPi-dependent phosphofructokinase (PP-PFK-1), that catalyzes the reverse reaction of F6P → F1,6BP, in regulatory steps that are similar to ones found in glycolysis and gluconeogenesis.
- fructose 1,6-bisphosphatase (FBPase-1) is inhibited by the allosteric modulator fructose 2,6-bisphosphate (F2,6BP)
- PPi-dependent phosphofructokinase (PP-PFK-1) is activated by fructose 2,6-bisphosphate
The coordinate regulation of these two enzymes is shown in Figure \(\PageIndex{4}\).
Figure \(\PageIndex{4}\): Regulation of sucrose synthesis at the formation of fructose-6-phosphate
The substrate for the PPi-dependent phosphofructokinase (PP-PFK-1) is PPi, which serves as a phosphodonor. In the plant cytosol, there is no pyrophosphatase to catalyze the cleavage of PPi into Pi. Also note that Fru-2,6-P2 itself is synthesized and degraded by the bifunctional enzyme phosphofructokinase 2/fructose-2,6-bisphosphatase, which we studied before in the regulation of the same step in glycolysis/gluconeogenesis.
The levels of F2,6-BP depend on the rate of photosynthesis:
- When photosynthesis is high (in light conditions), [DHAP] and [3PG] increase, which inhibits PFK2, which decreases F26BP, which causes a differential increase in F1,6BPase activity over PP-PFK-1), which increases F6P for sucrose synthesis as well as Pi for the continuation of the light reactions. This allows sucrose synthesis when excess DHAP and 3PG occur in the light reactions, which makes great biological sense.
- When photosynthesis is low (in dark conditions), the same regulations lead to an increase in F2,6BP, which leads to the preferential activation of the glycolytic enzyme PPi-dependent phosphofructokinase-1 (PP-PFK1) and inhibition of the gluconeogenic enzyme fructose 1,6- bisphosphatase (FBPase-1)
We will see later, the main regulatory step in starch synthesis is ADP-glucose pyrophosphorylase. In contrast to the inhibition by 3PG of PFK2, 3PG (which increases in active photosynthesis) activates ADP-glucose pyrophosphorylase while Pi inhibits it. Pi increases when the synthesis of ATP from ADP and Pi (by ATP synthase in the light reaction) slows (such as in darker conditions). If sucrose synthesis slows and sufficient 3PG persists, the activation of ADP-glucose pyrophosphorylase stimulates starch synthesis.
Glycosyltransferases (GTs)
Glycosyltransferases are very important enzymes as they are involved in the synthesis of most of the biomass on the planet. They catalyze the transfer of a monosaccharide from a donor that has been activated by the attachment of a nucleotide in the form of a nucleotide sugar (NDP-sugar) or dolichol-(pyro)phosphate sugar to acceptors. These include other sugars, lipids, and even proteins, which get glycosylated on alcoholic side chains (Ser, Thr) or amides (Asn). There are over 500,000 different GTs known and deposited in the Carbohydrate-Active enZYmes Database (CAZy database2). Based on sequence and structure there are over 114 different families. Although they depart significantly in primary sequence, only 3 major folds are predominant (GT-A, -B, and -C)
a glycosyl transferase reaction is required for the transfer of glucose from a donor like UDP-glucose to an acceptor like fructose to form sucrose as shown in the reaction below.
NDP-Glc (donor) + F6P (acceptor) → Sucrose-6-P + NDP
Perhaps now is a good time to study their generic reaction mechanisms before we move on to starch synthesis.
About 65% of glycosyltransferase reactions use nucleotide sugars as donors. These enzymes are called Leloir transferases. They are nucleotide-dependent. The activated NDP-sugar donor binds first to the enzyme, followed by the acceptor, to form a ternary complex. A conformational change allows catalysis to occur, which leads to the sequential release of products. The enzyme hence follows a sequential ordered bi-bi mechanism.
The reaction could proceed with either retention or inversion of the anomeric carbon of the donor NDPsugar. This is illustrated for the reaction of a C1 α-NDP donor monosaccharide with a monosaccharide acceptor to produce the α(1,4) link with retention of configuration or the β(1,4) link with inversion as shown in Figure \(\PageIndex{5}\).
Figure \(\PageIndex{5}\): Reaction of a donor NDP-monosaccharide and an acceptor monosaccharide with retention or inversion of configuration at the anomeric carbon of the donor
The same stereochemical outcomes can occur in the hydrolysis of acetal bonds by glycosyl hydrolases.
Reactions that proceed with inversion react in an SN2 reaction, similar to the nucleophilic attack on alkyl halides. For the glycosyl transferase that proceeds with inversion, the attacking nucleophile on the acceptor is made more nucleophilic by general base catalysis by a deprotonated glutamic or aspartic acid.
The glycosyl transferase that proceeds with the retention of configuration is less understood. Several alternative mechanisms have been proposed for both inverting and retaining glycosyltransferase in general, as illustrated in Figure \(\PageIndex{6}\).
Figure \(\PageIndex{6}\): .Proposed glycosyltransferase mechanisms. Schuman et al. PLOS One (2013). doi:10.1371/journal.pone.0071077.g001. Creative Commons Attribution License
They include the following possible mechanisms:
Panel (A): A double displacement mechanism utilizing two inversions with net retention of stereochemistry involving a covalent glycosyl-enzyme intermediate. The individual steps are inverting via (B) an SN2 process.
Panel (B): Inverting Leloir glycosyltransferases promote a backside nucleophilic attack on C1 by the acceptor from an inline (usually equatorial) position, resulting in inversion of the anomeric bond stereochemistry.
Panel (C): An orthogonal mechanism consisting of nucleophilic attack on C1 by the acceptor concurrent with leaving group loss from a position approximately at right angles to the C1-leaving group axis.
Panel (D): An SNi mechanism involving an intermediate with oxocarbenium character followed by rapid internal nucleophilic attach by the acceptor nucleophile; or
Panel (E): An SN1 mechanism involving a discrete oxocarbenium intermediate. All mechanisms require proton transfers of the hydroxyl hydrogen of the acceptor to an enzymatic base or the departing leaving group
You will remember from your studies of chemistry that SN1 reactions are dissociative and form a positively charged carbocation intermediate. In a SNi, reaction, the intermediate has cation character but the intermediate is not fully charged. In the case of glycosyltransferase, the intermediates would be oxycarbeniums. (Carbenium ion can be considered carbocations with 3 bonds to the carbon). SN1 reaction will occur only if the formation of the ion is activated and they are stabilized. A protic solvent is typically required for stabilization if the reaction occurs in solution. In the anhydrous active site of the enzyme, an appropriate arrangement of backbone and side chain negative or partially negative atoms is required to provide stabilization for SN1 and SNi mechanisms.
The double replacement reactions (Panel A) require a side chain nucleophile and likely candidates in retaining glycosyltransferase are not positioned for such a task. The evidence seems to support the orthogonal mechanism. It appears that the binding of the donor is similar in retaining transferase such that it is in a 900 position of nucleophilic attack by the acceptor, which leads o a trigonal bipyramidal transition state with the nucleophile axial the leaving group equatorial (orthogonal).
The donor NDP-monosaccharides typically are Mn2-containing proteins with the inverting and retaining transferase having different coordination geometries for Mn2+ binding, as illustrated in Figure \(\PageIndex{7}\).
Figure \(\PageIndex{7}\): Coordination geometries for Mn2+ binding in glycosyltransferase. Shuman, ibid.
Inverting enzymes such as GalT1 (top) achieve nearly perfect octahedral geometry about the coordinated metal ion (displayed angles of 81° and 91° compared to ideal octahedral 90° bond angles) with subsequent “inline” (approaching 180°) placement of the acceptor nucleophile for classic inverting SN2 backside attack. Retaining enzymes such as GTA (bottom), however, use an arrangement of acidic residues, often with acute bidentate Asp coordination, which severely skews metal geometry (displayed angles of 54° and 115°) and allows sufficient room between phosphate oxygens for orthogonal attack from the acceptor. Uis uridine, C1is donor galactose C1.
Figure \(\PageIndex{8}\) shows a possible mechanism for the transfer of a monosaccharide from the donor ADP-sugar through an oxycarbenium intermediate to an acceptor.
Figure \(\PageIndex{8}\): Possible mechanism for the transfer of a monosaccharide from the donor ADP-sugar through an oxycarbenium intermediate to an acceptor (example - a growing starch chain). Schuman B, Evans SV, Fyles TM (2013) Geometric Attributes of Retaining Glycosyltransferase Enzymes Favor an Orthogonal Mechanism. PLoS ONE 8(8): e71077. doi:10.1371/journal.pone.0071077. Creative Commons Attribution License
As mentioned above, there are three major folds for glycosyltransferases, GT-A, GT-B, and GT-C. Different representations of the structure of the GT-A fold core predicted through analysis by neural networks and deep learning are shown in Figure \(\PageIndex{9}\).
Figure \(\PageIndex{9}\): Fold core of GT-As. Taujale, R., Zhou, Z., Yeung, W. et al. Mapping the glycosyltransferase fold landscape using interpretable deep learning. Nat Commun 12, 5656 (2021). https://doi.org/10.1038/s41467-021-25975-9. Creative Commons Attribution 4.0 International License. http://creativecommons.org/licenses/by/4.0/.
This computational modeling of structure uses simple secondary structure representations generated from primary sequences to predict folds irrespective of sequence. GT folds are predicted with high accuracy by learning secondary structure features free of primary sequence alignments.
Panel (a) shows a linear map of the conserved secondary core structures (below) and graphs (top). The blue line represents a conservation score and the red a CAM score. CAM values correspond to residue positions that distinguish them the most from other class labels (folds and families).
Panel (b) shows a structural alignment of the core with the CAM values mapped onto it. The conserved regions are shown to have a high CAM value indicated by a high intensity of green and a low CAM value of purple.
Panel (c) shows on the left the consensus secondary structure for the aligned positions in the two GT-A fold clusters (blue: beta-sheets; red: helices; green: loops). Average CAM values from using different "layers" of analysis are shown for each aligned position (higher intensity of green corresponds to a higher CAM value). Cyan and magenta boxes highlight the secondary structure differences between the two clusters near the hypervariable HV2 and HV3 region respectively. The conserved DXD motif, G-loop, and C-His are indicated for reference. Donor and acceptor substrates for GT-A0 are shown as sticks
This figure is shown to give readers a sense of the complexity of the analysis required to understand and differentiate structure/function features for these structurally similar but complex enzymes.
Structure and Enzymatic Activity of Sucrose Synthase (SuSy)
These enzymes are usually homotetramers with a monomeric molecular weight of around 90,000. The monomers typically have an N-terminal domain that directs the targeting of the enzyme to a specific location and a C-terminal GT-B domain. It can be regulated by phosphorylation at a serine near 12 in the N-terminal domain, which presumably regulates its cellular location, and other near 170 that affects its degradation. Two glutamates in the C-terminal GT-B domain (E678 and E686) and phenylalanine (680) are essential for the catalytic activity. Sucrose synthase is reversible as is the synthesis of sucrose-6-P from F6P and UDP-glucose can be reversed in the presence of UDP. The enzyme can also use ADP-glucose as a donor.
The structure of the Thermosynechococcus elongatus sucrose phosphate synthase with bound UDP and sucrose-6-Phosphate has been solved and along with other studies a reaction mechanism proposed as shown in Figure \(\PageIndex{10}\).
Figure \(\PageIndex{10}\): Catalytic model of TeSPS. Yuying et al. Co-crystal Structure of Thermosynechococcus elongatus Sucrose Phosphate Synthase With UDP and Sucrose-6-Phosphate Provides Insight Into Its Mechanism of Action Involving an Oxocarbenium Ion and the Glycosidic Bond. Frontiers in Microbiology, 11, 2020. https://www.frontiersin.org/article/...icb.2020.01050. Creative Commons Attribution License (CC BY).
(A)The state before the reaction is shown. (B)The glucose residue of UDPG forms hydrogen bonds between/among the phosphate groups, His158, Glu331, and F6P. Due to the formation of these hydrogen bonds, the pyranose ring of the glucose becomes negatively charged to promote C1 to form an oxocarbenium ion. (C)The relatively weak hydrogen bond formed by His158 and O6 is broken, which causes the pyranose ring to lose some negative charge character and forces the C1 oxocarbenium ion to form a covalent bond with the F6P oxygen atom. (D)UDP and S6P are released from the catalytic center.
A 2D view of the active site residues is shown in Figure \(\PageIndex{11}\).
Figure \(\PageIndex{11}\). The catalytic center of TeSPS. Yuying et al. ibid.
The catalytic center of TeSPS. Loops 1, 3, 4, and 5 forms a cave that binds to the uracil moiety of UDP. Glu339 stabilizes the ribose ring via the formation of two hydrogen bonds. Leu335 forces two phosphate groups in UDP to reorient. Several basic amino acids, including Arg105, Arg178, Arg249, and Arg253, interact with the phosphate groups of UDP and S6P via ionic bonds. Pro332 at the turn of loop 6 interacts with the pyranose ring via CH/π bonds. All hydroxyl groups (O2, O3, O4, and O6) of the glucose moiety of S6P form hydrogen bonds with phosphate groups or the side chains of various amino acids. “O2” forms a hydrogen bond with P1O1 of the P1 phosphate group of UDP. “O3” forms a hydrogen bond with the carboxyl group of Glu331. “O4” forms a hydrogen bond with P2O1 of the P2 phosphate group of UDP. “O6” forms a hydrogen bond with the imidazole side chain of His158. The distances between groups are indicated in the figure
Figure \(\PageIndex{12}\) shows an interactive iCn3D model of the Thermosynechococcus elongatus Sucrose Phosphate Synthase With UDP and Sucrose-6-Phosphate (6KIH)
Figure \(\PageIndex{12}\): Thermosynechococcus elongatus Sucrose Phosphate Synthase With UDP and Sucrose-6-Phosphate (6KIH) (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...1kCnQ9pxcJjdy7
Metabolism of Sucrose
Sucrose is a primary synthesis product of photosynthesis and is transported to other plant "sink" tissues where it is used for both energy and biosynthetic precursors. Suc transporters can move it from the phloem to the apoplast. It can enter sink cells through Suc transporters or be hydrolyzed by cell-wall invertase (cwINV) to yield glucose (Glc) and fructose (Fru), which enter by hexose transporters. Suc can also pass directly from the phloem to sink cells through plasmodesmata (physical connections between cells). Inside sink cells, Suc can be metabolized or transported to the vacuole, where it can be stored as Suc, transformed into fructans by fructosyltransferases (FTs), or hydrolyzed by vacuolar invertase (vINV) and stored as hexoses. To be metabolized, Suc must be hydrolyzed by either cytosolic invertase (INV) into glucose and fructose, or by the reversible reaction of sucrose synthase (SuSy) using UDP instead of water to yield fructose and UDP-G. These processes are illustrated in Figure \(\PageIndex{13}\).
Figure \(\PageIndex{13}\): Simplified schematic presentation of sugar metabolism in sink tissue cells toward cellulose, callose, and starch synthesis. Stein and Granot. Frontiers in Plant Science (10). 2019. https://www.frontiersin.org/article/...pls.2019.00095. DOI=10.3389/fpls.2019.00095 https://www.frontiersin.org/articles...019.00095/full
The hexoses (glucose and fructose) can be phosphorylated to hexose phosphates (hex-P), directed to starch synthesis in the plastid or glycolysis, and then respiration in the mitochondria or directed to other metabolic pathways. Plasma membrane-associated SuSy (pmSuSy) and cwSUS can generate UDP-G that is used in the synthesis of cellulose for cell walls and callose for plugging plasmodesmata. Callose is a polysaccharide in the form of β-1,3-glucan with some β-1,6-branches in the cell walls of a wide variety of higher plants
Biosynthesis of Starch
During active photosynthesis in bright light, a plant leaf produces more carbohydrates (as triose phosphates) than it needs for generating energy or synthesizing precursors. The excess is converted to sucrose and transported to other parts of the plant, to be used as fuel or stored. In most plants, starch is the main storage form, but in a few plants, such as sugar beet and sugarcane, sucrose is the primary storage form. The synthesis of sucrose and starch occurs in different cellular compartments (cytosol and plastids, respectively), and these processes are coordinated by a variety of regulatory mechanisms that respond to changes in light level and photosynthetic rate.
Starch Synthesis
Starch, like glycogen, is a homopolymer of D-glucose in (α1,4) linkage with (α1,6) branches. Glycogen is found in Archaea, Bacteria, and Eukaryotes. In contrast, starch is found in photosynthetic algae, land plants, and in some cyanobacterial species.
Starch is synthesized by starch synthase in chloroplasts for temporary storage and in amyloplasts in seeds, roots, and underground stems (tubers) for long-term storage. As with the synthesis of glycogen, the glycosyltransferase catalyzes the addition of an activated ADP-glucose to the acceptor, the elongating starch polymer.
ADP-α-D-glucose-1-phosphate (donor) + [(1→4)-α-D-glucosyl](n) (acceptor) ↔ [(1→4)-α-D-glucosyl](n+1) + ADP + H+
The ADP-glucose donor is formed in the following reaction:
Glucose 1-phosphate + ATP ↔ ADP-glucose + PPi
In plastids, there is, in contrast to the cytosol, a pyrophosphatase which makes the reaction irreversible. Hence the next overall reaction is
Starchn + glucose 1-phosphate + ATP → starchn + 1 + ADP + 2Pi
Taking into account the hydrolysis by inorganic pyrophosphatase of the PPi produced during ADP-glucose synthesis, the overall reaction for starch formation from glucose 1-phosphate is:
Starchn + glucose 1-phosphate + ATP → starchn + 1 + ADP + 2Pi; ΔG'º = -50 kJ/mol = -12 kcal/mole (-50 kJ/mol)
ΔG'º = -50 kJ/mol = -12 kcal/mol
In glycogen synthesis, the donor is UDP-glucose, and it is added to the reducing end (C1) of the growing glycogen polymer (with the C1 OH acting as a nucleophile) so the polymer extends from that end. Kinetic models suggest that starch synthases, which use ADP-glucose, may use two different active sites that appear to alternately add glucose to the nonreducing C4 end (with the C4 OH acting as a nucleophile), with the reducing end of the linear α(1,4) polymer being alternately covalently attached to one site, then the other, with the attachment activating that end for reaction with C4-OH of the polymer at the other site.
Figure \(\PageIndex{14}\) shows a very simplified structure of the starch synthase using two different active sites as the reaction proceeds through the first steps.
Figure \(\PageIndex{14}\) shows a simplified structure of the starch synthase using two active sites
Note that additional glucose units are added to the nonreducing C4 end denoted by a star. α(1,6) branches are added by branching enzymes as in the case of glycogen. Bacteria starch is made in a fashion similar to glycogen, but they use ADP-glucose as do plant starch synthase. Structural models show that the protein has one active site so the kinetic models suggesting the use of two active sites may refer to a movement of chains between different monomers in oligomeric forms of the protein.
Both the donor (ADP-Glc) and product (starchn+1) have their participating electrophile (ADP) and now substituted nucleophile (C4-O-R) in the α-anomeric form so they are retaining glycosyltransferases. The enzymes could act in two different ways:
- processive mechanism: the acceptor (starchn) stays bound to the enzyme after each addition of the next glucose from the donor ADP-glc, and one chain extends quickly
- distributive mechanism: the enzyme dissociates from the product (starchn+1) after the addition of the glucose from ADP-glc, and must rebind to catalyze the next addition, so many new chains start and the growth of each chain is slow
Kinetic evidence suggests that some starch synthases are processive and others are distributive. Along with starch synthase, three other enzymes are involved, ADP-glucose pyrophosphorylase (AGPase), starch branching enzyme (SBE), and starch debranching enzyme (DBE)
Starch synthesis is regulated by gene transcription, phosphorylation, and redox conditions. A key regulatory enzyme is an ADP-glucose pyrophosphorylase (AGPase). This enzyme catalyzes the formation of ADP-glucose (the donor) and PPi from glucose 1-phosphate and ATP. Dithiothreitol, a reducing agent, increases starch synthesis by inactivating AGPase. We will see below that a key disulfide bond is some starch synthases that must be reduced (cleaved) to open an active site cleft between the N-terminal and C-terminal lobes of the catalytic domain. In vivo, thioredoxins are probably involved in redox regulation.
Structures of starch synthetases (SS)
The are four classes of soluble starch synthetases (SSI-SSIV) and one starch granule-bound one (GBSS). All have two catalytic domains as noted in the mechanism above except SSII which has 3 CHO binding domains. GBSS appears to form amylose and long chains of amylopectins (amylose with around 5% α(1,6) branching. Loss of function mutants of GBSS have much-reduced amylose concentrations. SSI-SSIII produces most of the amylopectin. SSI is most active with stands (outer ones in branched structures) that have around 7-9 glucoses. In leaves, it generates chains of up to about 10 residues in amylopectin. The SSs can be chloroplastic or amyloplastic.
Figure \(\PageIndex{15}\) shows an interactive iCn3D model of barley starch synthase I in complex with maltooligosaccharide (4HLN).
Figure \(\PageIndex{15}\): Barley starch synthase I in complex with maltooligosaccharide (4HLN). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...LGwFUeJAMskBcA
The protein is shown in gray in an open conformation a glucose 5-mer bound on the outside. The protein has a GT-B fold with distinct N- and C-terminal Rossmann-like domains and a central linker. Side chains within 5 Å of oligosaccharide are shown in sticks and labeled. Note especially the disulfide bridge between cysteines 126 and 506 in the central part of the protein that prevents the formation of the active site. This clearly shows the importance of redox signaling to activate the enzyme.
The maltose is not bound in the active site but at a surface secondary binding site (SBS). The role of this site is a bit unclear but may be involved in carbohydrate:carbohydrate interactions. Specifically, they may assist in recruiting starch chains for further elongation. SBSs are found in many but not all starch synthases. Note that in this structure there is only one occluded active site and not two as suggested in Figure 14 above.
Figure \(\PageIndex{16}\) shows an interactive iCn3D model of the catalytic domain of starch synthase IV from Arabidopsis thaliana bound to ADP and acarbose (6GNE).
Figure \(\PageIndex{16}\): Catalytic domain of starch synthase IV from Arabidopsis thaliana bound to ADP and acarbose (6GNE) https://structure.ncbi.nlm.nih.gov/i...MioUaKM73M3Wd9
This structure is just the catalytic domain (representing about half of the total protein sequence). The N-terminal domain is colored by the secondary structure. Acarbose (spacefill, CPK colors) again occupies both the donor and acceptor sites in the active site (central regions). The structure has a secondary binding site (SBS) occupied by the disaccharide maltose. Again there are not two active sites, but the migration of starch chains between monomers in oligomeric forms could support the model shown in Figure 14.
The structure of beta-acarbose, an inhibitor, is shown in Figure \(\PageIndex{17}\).
Figure \(\PageIndex{x18}\) below shows an interactive iCn3D model of the Granule Bound Starch Synthase from Cyanobacterium sp. CLg1 bound to acarbose and ADP (6GNF)
Figure \(\PageIndex{18}\): Granule Bound Starch Synthase from Cyanobacterium sp. CLg1 bound to beta-acarbose and ADP (6GNF). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?F1tJBDAh6QRhxpH88
ADP, occupying part of the donor site that would usually bind NTP-Glc, is shown in CPK-colored sticks. The rest of the donor site (where Glc of NTP-Glc would bind) and the acceptor site (growing starch chain) is occupied by beta-acarbose, It is shown in the CPK-colored wire-frame surface. Histidine 181, which probably stabilizes an " oxocarbenium-like anomeric carbon in the transition state", is shown in ball and sticks with CPK colors.
Cellulose Synthesis
We can't leave plant carbohydrate metabolism without considering the synthesis of cellulose, which is catalyzed in plants by members of the superfamily cellulose synthase (CesA) and cellulose synthase-like (CsI) enzymes, which are part of the glycosyltransferase GT2 family and have similar structures. Cellulose and hemicellulose are, of course, chief components of the 10 and 20 cell walls. Members of the CesA family have a conserved motif (DDDQxxRW) as well as a zinc-finger domain. Different members catalyze the synthesis of the 10 and 20 cell walls. Members of the Csl family are involved in additional cell wall glycans including (1,4)-β-D-mannan (CsIA) and xyloglucan cytoskeleton (CslC). UDP-glucose is the donor in the creation of the β(1,4) acetal linkages between glucose monomers.
Plant growth must respond to environmental triggers through a balance of cell expansion and cell division and a key regulator of these processes is the flexibility of the cell walls which can maintain turgor pressure by expansion. Cells that are nonexpanding (for example those that line the xylem vessels and in woody tissue) have secondary cell walls beneath their primary walls.
We have previously discussed the structure of the primary and secondary cell walls in Chapter 7.3. In brief, the primary cell walls contain cellulose, hemicellulose, and pectins. Cellulose, the main component that provides strength, is synthesized by CesA which forms a very large rosette-shaped complex (CSC). These complexes seem to move intracellular microtubules which guide that synthase complex through the interactions of microtubule-associated cellulose synthase compartments (MASCs), whose numbers increase during stress. Likewise, there are uncoupling proteins that inhibit microtubule movement from the CSC. These protein complexes stay aligned during cell growth. The cell wall hence is a key player in signal transduction that allows growth and cell division.
The structure of the rosette-shaped complex (CSC) has been determined by cyroEM and is shown in Figure \(\PageIndex{19}\).
Figure \(\PageIndex{19}\): Structural cartoons of the CESA CSC complex. Nixon, B., Mansouri, K., Singh, A., et al. Comparative Structural and Computational Analysis Supports Eighteen Cellulose Synthases in the Plant Cellulose Synthesis Complex. Sci Rep 6, 28696 (2016). https://doi.org/10.1038/srep28696. Creative Commons Attribution 4.0 International License. http://creativecommons.org/licenses/by/4.0/
Panel (A) shows the complex as a series of trimers of CESAs, with each monomer spanning the membrane with 7 alpha helices. The catalytic domain is in the cytoplasm.
Panel (B) shows the CSC complex with the top leaflet removed
Panel (C) shows a top-down view showing 6 sets of trimers of CESA.
Panel (D) shows a metal replica viewed in the TEM after the removal of the biological material.
Each trimer synthesizes a cellulose strand. There are 18 trimers in the complex allowing the concomitant synthesis of cellulose strands that can easily self-associate through hydrogen bonding to form near the extracellular surface cellulose protofibrils.Figure \(\PageIndex{20}\) shows an interactive iCn3D model of the catalytically active homotrimeric poplar cellulose synthase (6WLB)
Figure \(\PageIndex{20}\): Catalytically active homotrimeric poplar cellulose synthase (6WLB). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...zCm6fYSwAFA897
Each of the monomers within the trimers is given a different color. A 5 residue β(1,4) glycan is shown in cartoon form emerging into the middle of the membrane complex.
Figure \(\PageIndex{21}\) shows an interactive iCn3D model of the homotrimeric poplar cellulose synthase isoform glycan binding site (6WLB).
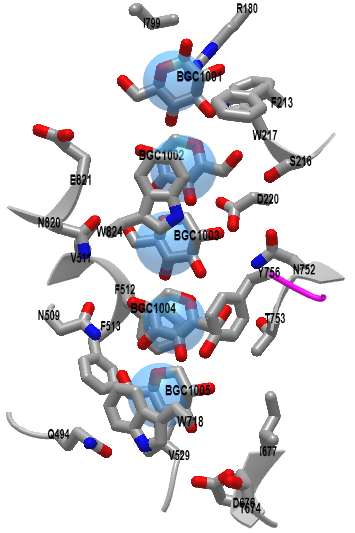
Figure \(\PageIndex{21}\): Homotrimeric poplar cellulose synthase isoform glycan binding site (6WLB. (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...HqYhTQfuw857PA
The actual transmembrane channels start just above the active site. Key amino acid side chains (Trp 718, Phe 513, Val 529, and Gln 494) help form the portal opening. The actual channel is lined with both aromatic and hydrophilic residues, which supply sufficient but not too strong noncovalent interactions that allow sequential movement of the continually-synthesized cellulose as it ratchets forward toward the extracellular side of the membrane. The aromatic residues interact through pi stacking with the glucose residues and through interactions with equatorial OH groups on the β-glucose polymer. Remember that cellulose is especially stable from a steric perspective since all its OH groups and the acetal linkage are equatorial.




.png?revision=1&size=bestfit&width=413&height=272)
.png?revision=1&size=bestfit&width=380&height=272)
.png?revision=1&size=bestfit&width=324&height=218)
.png?revision=1&size=bestfit&width=383&height=287)
.png?revision=1&size=bestfit&width=448&height=323)