11.4: Diffusion Across a Membrane - Pores
- Page ID
- 108139
Pores and Pore-Forming Proteins (PFPs)
If you form a pore in a cell bilayer, molecules of all sizes could move either way based on their electrochemical potential. They will move from regions of a higher to lower electrochemical potential in a thermodynamically favorable process. Hence movement through pores represents a special case of facilitated diffusion when part of the driving force is not just a concentration gradient but also an electrical potential. Several questions might come to mind.
- What proteins are involved in pore formation?
- How is the specificity of solute movement through the pore regulated?
- What is the mechanism of pore formation?
Pore formation can lead to cell death, which is the function of some pore-forming proteins (PFPs) including the toxin Hemolysin E (also known as HlyE, ClyA, SheA) secreted from E. Coli and S. Aureus. Human proteins also form a membrane attack complex (examples include the membrane attack complex-perforin/cholesterol-dependent cytolysin (MACPF/CDC) superfamily and the membrane attack complex (MAC). The MAC consists of an assembly of proteins involved in the complement system (part of the effector branch in the innate immune system), which leads to cell death of Gram-negative bacteria like E. Coli. Figure \(\PageIndex{1}\) illustrates the assembly process of the membrane attack complex and the complexity of interactions required to form a lethal pore in a cell.
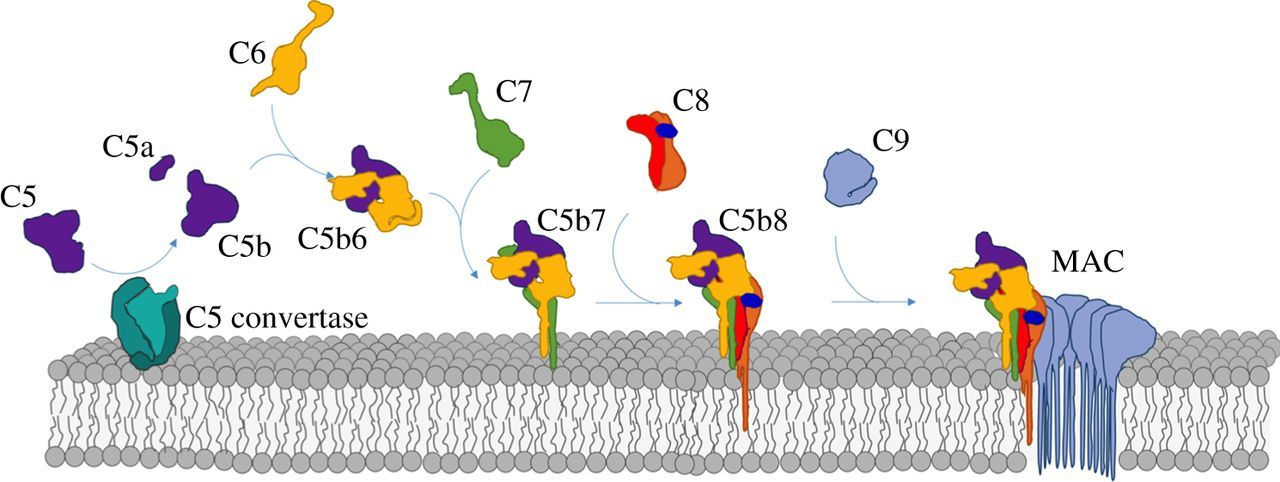
Complement protein C9 can adopt a soluble form or membrane form which in aggregates form the pore leading to cell death. This is a common feature of PFPs.
PFPs could create a pore by altering membrane lipid packing to form a toroid-like hole (Figure \(\PageIndex{2}\)) and/or by inserting in a membrane and forming a pore within the protein complex itself. In either mechanism, lipid packing is altered.
Biophysical evidence shows some support for the formation of "toroidal pores".
Lipid rearrangements in the membrane could lead to a hydrophilic or hydrophobic pore lining as shown in Figure \(\PageIndex{3}\).
We started our study of lipid bilayers with pure lipid systems and then added membrane proteins. Let's do the same with pore formation. A common technique to form a pore in pure lipid bilayers and also in cells is electroporation. This is a technique used to move a DNA with a target gene into either a prokaryotic cell (transformation) or eukaryotic cell (transfection) for exogenous gene expression. In the absence of PFPs, this requires the alteration of surface tension by applying an electrical potential. This forms depressions in the membrane, altering the nonpolar acyl chain packing. In the process, small wire-like columns of water appear, which like hydrophobic pores ultimately rearrange into hydrophilic toroidal pores. Figure \(\PageIndex{4}\) shows snapshots of molecular dynamics simulations as a function of the time of pore formation in electroporation.
How does DNA pass through the pores in the bilayer? In pure lipid vesicles, it appears to pass through by electrophoresis. Most students are familiar with the electrophoresis of DNA fragments through pores in agarose gels. In actual living cells, small nucleic acids like small interfering RNA (siRNA) and antisense DNA molecules appear to pass through the bilayer by electrophoresis. Large DNAs like plasmids containing a gene for expression bind to the cell and form cell surface aggregates, which appear to be endocytosed into the cell. Electroosmosis, the movement of liquids under the influence of an electric field, also plays a role.
Pores - Outer Membrane Factor (OMF) and Voltage-Dependent Anion Channel (VDAC)
Now let's consider pores made of PFPs. We have already discussed two types of beta-barrel transmembrane proteins, the outer membrane factor (OMF) of Gram-negative bacteria and the voltage-dependent anion channel (VDAC). Both are examples of proteins called porins with typical beta-barrel topology.
VDAC: At low membrane potentials, VDAC (also known as mitochondrial porin), the most abundant protein in the mitochondrial outer membrane, moves metabolites and Ca2+ ions across the outer membrane of mitochondria. VDAC exist in an open state at 0 or very low transmembrane potentials that allows for the transfer of key metabolic anions (pyruvate, oxaloacetate, malate, succinate, ATP, ADP, and Pi, which are involved in metabolism) and Ca2+, and in a closed state (above or below + 30 mV), which is not completely closed as it allows for the transfer of ions with a preference for cations. The closed state does not allow for the transfer of ATP. The transition to the closed state is promoted by tubulin and actin (cytoskeletal proteins), negatively charged lipids such as phosphatidyl ethanolamine and cardiolipin, and also covalent phosphorylation by protein kinases. Bcl2, proteins involved in the regulation of programmed cell death (apoptosis) also interact with VDAC. In contrast to ligand-gated channels which require ligand binding to open the channel, pore complexes are unusually open. Millions of ATPs/second move across the membrane through the open pore but none through the closed pore.
Figure \(\PageIndex{5}\) shows an interactive iCn3D model of mouse VDAC1 (4c69) with bound ATP loosely held in the site. An alpha helix partly occludes the central pore of this β-barrel protein.
.png?revision=1&size=bestfit&width=327&height=285)
One ATP is bound in the barrel and interacts with Lys 12 and Lys 20 at each end of the cavity-bound helix. The alpha-helix narrows the pore opening and presumably changes orientation in a voltage-sensitive fashion to gate the pore open and closed, hence regulating the conductance of ions through the pore. That the orientation of the charged arginine side chains would be dependent on the transmembrane potential should be somewhat obvious.
Aquaporins can move a billion water molecules per second across membranes and exclude ions including protons. Waters proceed in a single file through the pore. Instead of moving waters through, it might simply move "H+ ions" through by the alignment of subsequent H bond donors and acceptors in a "wire" of water molecules. This is prevented by two conserved asparagine residues in the center of the channel, which disrupt water to water hydrogen bond network in the channel waters that could facilitate proton transfer. Instead, the central waters form hydrogen bonds to the central asparagines. This, along with local membrane potentials, causes opposite orientations of the water in different leaflet sides of the membrane, precluding H+ transfer.
Here is a movie of a molecular dynamics simulation of water moving through a porin called aquaporin, GlpF, from E. Coli.
Science magazine (Tajkhorshid et al., Science Apr 2002, 296:525). Used with permission from the Theoretical and Computational Biophysics Group, the National Institutes of Health (NIH) Resource for Macromolecular Modeling and Bioinformatics, at the Beckman Institute, University of Illinois at Urbana-Champaign.
OMF: The outer membrane factor (see previous section) is one member of a class of bacterial porins, which are the most abundant proteins in the outer membrane of Gram-negative bacteria. They are classified as non-specific or specific (with respect to the solute that passes through), or monomeric, dimeric, or trimeric based on their structure. In Gram-negative bacteria, which have two lipid bilayers, the movement of solute from inside to outside includes at least three sets of proteins. Active transport (discussed in the next section) needs an energy source and is used by inner membrane transport proteins, including ATP-binding cassette (ABC)-type, resistance nodulation division (RND)-type and major facilitator superfamily (MFS)-type transporters. These are connected to membrane fusion proteins (MFP) that span the periplasm, which then interacts with at least 21 different types of porins. Molecules with molecular weights greater than 600 generally can not get through the nuclear envelope of Gram-negative bacteria, limiting the size of potential antibiotics, which must enter by passive diffusion.
Mechanosensitive ion channels - Mscs (which are pores!)
As the name applies, these ion channels (with openings large enough to be called pores) are gated open/closed by mechanical (physical) changes in the properties of the membrane, not extracellular/intracellular ligands or voltage changes. Certain bilayer lipids also activate the Mscs. There are two types, small (MscS) and large (MscL) mechanosensitive ion channels. Changes in local (boundary layer) and nonlocal lipids are involved in the gating of the channel (in the next section we will discuss lipid-gated ion channels). They are found in prokaryotes, archaea ,and eukaryotes. They are also called stretch-gated ion channels.
Mcss transduces a physical force (stretching and change in turgor pressure) into an electric signal - a flow of ions across the membrane. Turgor pressure is the internal pressure that "presses" the cytoplasm and cell membrane towards the cell wall in bacteria and plant cells. It arises mostly from the osmotic flow of water into the cell. If bacterial cells are placed in a high salt concentration solution (hypertonic), water flows out of the cell and the cell membrane shrinks to the inside of the volume confined by the cell wall. When placed in a hypotonic solution, water flows into the cell and the cell membrane presses out to the cell wall. The response of these channels is fast, in the millisecond range, which is about as quick as a cellular response can be. Some variants of these channels are called piezochannels, based on the piezoelectric effect that describes how a voltage is produced when some materials are deformed by mechanical stress which causes a redistribution of charges.
Bacteria normally have high concentrations of both K+ and negatively charged anions, especially glutamate, which leads to a high turgor pressure from the inward osmotic flow of water. At low external osmolarity, turgor pressure in the cell could be as high as 4 atm. If placed in external solutions of high osmolarity, bacterial cells respond by the increased movement of solutes into the cell.
Mscs are particularly important when bacteria are subjected to sudden osmotic shock. If they are placed in pure water, for example, water would flow down a concentration gradient into the cell and cause the cell to swell and lyse, killing the cell. This is done in the lab to prepare almost pure hemoglobin from ruptured red blood cells. The Mscs are opened under these conditions and small species from the cytoplasm flow out, helping to keep the cell viable. Their openings must be regulated to prevent too much outward flow, which would kill the cell.
In other organisms they are also involved in touch (stretch), hearing (vibration, sound waves), and responses to gravity. Stimuli to activate them include fluid shear stress (relevant to endothelial cells that line blood vessels), membrane stretch (relevant to skeletal and cardiac muscle cells), or even indentation of a bilayer with a pipette. Changes in transmembrane turgor or other mechanical pressures cause membrane tension. Yet even in the absence of these changes, Mscs can be activated by anesthetics, phospholipids missing one fatty acid (called lysophospholipids), and certain polyunsaturated lipids. These stimuli also perturb the membrane bilayers.
Given the pore size, these proteins are less selective to ion flow compared to voltage-gate channels (see next section). Depending on the amino acids that line the pore, some Mscs would allow the preferential flow of anions while some allow cation flow.
Some somatosensory channels (i.e. not pore) proteins also respond to pressure. When open, these can be selective to specific ions like Na+ or K+. Examples include some variants of the Transient Receptor Potential (TRP) ion channels. Other membrane proteins can also be activated by physical force. but true Mscs have some key characteristics. If mutated or deleted, the mechanosensory response is removed. If added to a cell, a mechanosensory response results.
a. Small-conductance mechanosensitive channel
This protein is a homoheptamer with three helices from each monomer involved in the overall structure. Two (helices 1 and 2) interact more with the lipid components of the bilayer. Interactions of specific lipids with the helices seem to promote closing but changes under high pressure lead to the opening of the pore. Half of each helix 3 forms the pore, while the other half is more parallel to the membrane and interacts with a large cytoplasmic domain.
Figure \(\PageIndex{6}\) shows the differences between the closed form of E. Coli MscS ( 2oau) and the open form (2vv5) viewing down the pore axis. The heptameric protein is shown in gray. Two key valines (105 and 109) on each chain are shown in spacefill and colored cyan. These hydrophobic amino acids act like gate-keepers, helping to keep water out, acting like a "vapor seal". In the closed state, the pore is sealed by the closing of the leucine "rings" as one half of helix 3 pack more closely. Many members of the MscS family vary significantly in size and can have between 3-11 transmembrane regions. The closed pore has a diameter of about 4.8 Å, while the open pore is 13 Å across.

Most of you would have studied Ohm's law, given by
\begin{equation}
\mathrm{V}=\mathrm{IR} \text { or } \mathrm{I}=\frac{\mathrm{V}}{\mathrm{R}}
\end{equation}
where I (amps) is the current, R (ohms) is the resistance and V (volts) is the voltage. A more general variant of this law used in physics is
\begin{equation}
\mathrm{J}=\sigma \mathrm{E}
\end{equation}
where J is the current density, E is the electric field, and σ is the conductivity (inverse of resistance) which depends on the material. The unit of sigma σ is ohms-1 or mhos (ohm written backward). That unit has been renamed the Siemen (S). Mscs channels have a small conductance of approximately 1 nS in 400 mM salt solution.
b. Large conductance mechanosensitive ion channel (MscL):
The channels have large conductances (3 nS) and concomitantly larger pore sizes, allowing the flow of water, ions, and even small proteins. Again they are involved in diffusion down an electrochemical gradient through pores so not active transport just gated diffusion.
Figure \(\PageIndex{7}\) shows an interactive iCn3D model of the pentameric MscL from Mycobacterium tuberculosis (2oar), a Gram-positive bacteria that causes tuberculosis. About 23% of the world's population is affected by this pathogen. It causes about 1.5 million deaths each year. Compare this to the total number of deaths during the COVID pandemic (2020-21) of over 3 million (as of May 2021). It has killed over 1 billion people throughout human history (but not as many as malaria). The pore diameter is about 30 Å across.
.png?revision=1&size=bestfit&width=249&height=259)
Pore-forming alpha-helical toxins
We started this chapter section with a discussion of the major attack complex (MAC) of the innate immune system. Pathogens also employ pore formation to kill host cells. Many secrete soluble proteins which aggregate in the membrane to form either alpha-helical or beta-barrel pores. The proteins are called pore-forming toxins (PFTs). Killing occurs when either cytoplasmic components leak out or bacterial toxins, such as diphtheria and anthrax toxin, move into the cell.
Figure \(\PageIndex{8}\) shows an interactive iCn3D model of the pore formed by cytolysin A (ClyA, also known as HlyE), an alpha-PFT used by some E. Coi and Salmonella enterica strains. The pore is a large dodecamer that forms from soluble monomeric ClyA (2wcd). It has a pore diameter of about 40 Å,
_(2wcd)_.png?revision=1&size=bestfit&width=319&height=404)
Figure \(\PageIndex{9}\) shows the soluble monomeric form of cytolysin A (ClyA, HlyE) (1QOY). Nonpolar side chains are shown in cyan. The transparent surface is mostly polar, making the monomer soluble.
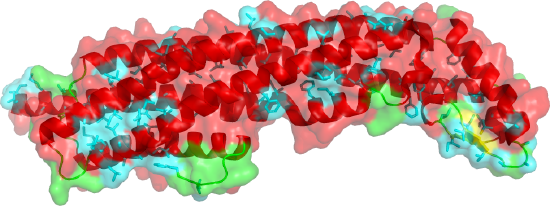
Figure \(\PageIndex{10}\) shows the changes in conformation between the single chain soluble form (shown in the figure above) and a single chain of the membrane oligomeric channel.

We started this chapter section by exploring electroporation and the formation of a toroidal-like hole in the lipid bilayer. In the case of ClyA, the protein aggregate itself forms the pore, not the lipids themselves, although lipid rearrangements are necessary to form the protein complex.
Gap Junctions
Connexins are voltage-gated channels that allow for the flow of ions, metabolites, nucleotides, and small peptides. A connexin has four transmembrane helices and two extracellular loops, Six protomers of these come together in a single cell to form a channel complex called a connexon or hemichannel. Beta structures in the connexin hemichannel of one cell docks with a similar channel on an adjoining cell to form a full channel passing through the membranes of both cells forming a gap junction between the cells. This is shown in Figure \(\PageIndex{11}\). There are 20 different connexins encoded in the human genome.
The left figure below shows the six protomers, each in a different color and a gray rectangle representing the bilayer of a single connexon or hemichannel. The right side of the figure shows a full gap junction channel between two cells, with the membranes represented by gray rectangles. The connexin 26 monomer was used to create the diagram. Mutations in this protein are associated with hearing loss.
The left figure above shows the six protomers, each in a different color and a gray rectangle representing the bilayer of a single connexon or hemichannel. The right side of the figure shows a full gap junction channel between two cells, with the membranes represented by gray rectangles. The connexin 26 monomer was used to create the diagram. Mutations in this protein are associated with hearing loss.
The cytoplasmic entrance of the channel is positively charged. It forms a funnel, composed of 6 amino-terminal helices, which leads into a negatively charged transmembrane lining. The entrance diameter is 14 Å.
Here is a model of a full gap junction channel connecting two membranes using the human connexin 26 monomer (2zw3). Only the bottom membrane bilayer is represented by red and blue dummy atoms.
Figure \(\PageIndex{12}\) shows an interactive iCn3D model of a full gap junction channel connecting two membranes using the human connexin 26 monomer (2zw3). Only the bottom membrane bilayer is represented by red and blue dummy atoms.
.png?revision=1&size=bestfit&width=246&height=329)
The Nuclear Pore Complex (NPC)
Channels have pores that can be gated open and allow the selective flow of ions. Pore-forming proteins have larger entrances that allow both small and large molecules to pass through the bilayer. The pore opening in even large mechanical sensitive channels (about pale in size compared to the nuclear pore complex, which has a combined molecular mass of around 125,000,000! Its outer diameter of ~1,200 Å and its inner one of about 425-Å. Figure \(\PageIndex{13}\) shows the relative size of nuclear pore compared to other molecular structures including the eukaryotic ribosome, nucleosome, a soluble tetrameric protein (rubisco, 270K), and MscL (shown as a circle which represents the pore diameter).
Its job is to shuttle small molecules by passive diffusion down a concentration gradient through the pore. In addition, it moves large molecules and molecular structures (proteins, RNA, and perhaps ribosomes) across the nuclear membrane in a process that requires energy. The proteins that comprise this complex are called nucleoporins (nups), of which there appear to be around 34 in humans. Each NPC complex contains around 1000 nucleoporins. The complex fuses the inner and outer nuclear membranes.
We have focused so much on single bilayer membranes that comprise the plasma membrane and membranes of organelles like the Golgi complex and lysosomes, it might come as a surprise (perhaps not to biology students) that the nuclear membrane or envelope appears to consist of two bilayers. Most know that the mitochondria have two bilayers, an inner and outer membrane, similar to Gram-negative bacteria. Mitochondrial are believed to have arisen from bacteria so the double bilayer there makes sense. Figure \(\PageIndex{14}\) shows the nuclear membrane or envelope of two bilayers (1) with an outer ring (2), spokes (3), a basket (4), and filaments (5). The NPC spans both bilayers.
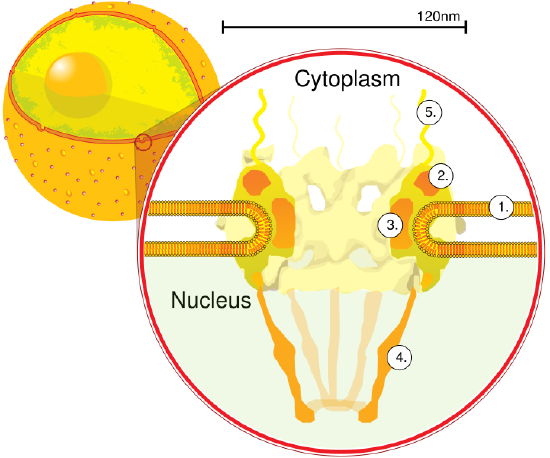
The outer bilayer of the nuclear envelope is continuous with the endoplasmic reticulum as shown in Figure \(\PageIndex{15\) below. The dots on the ER membrane are ribosomes, making this the rough ER (as opposed to smooth ER, which has no attached ribosomes).
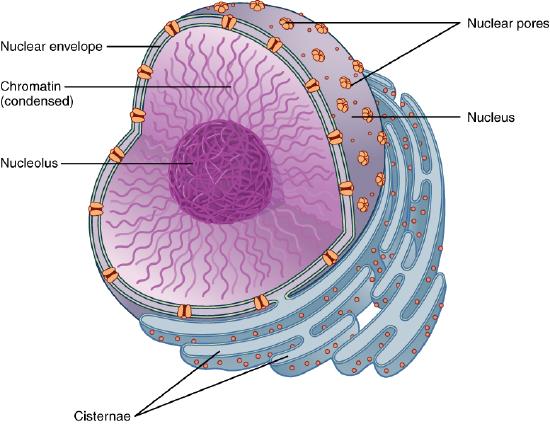
Figure \(\PageIndex{16\) below shows a model of the basket-like structure of the nuclear pore complex (NPC). It, as well as Figure XX, shows that instead of two separate bilayers, in actuality, there is just one bilayer with each leaflet bending around at the NPC and reversing directions! Think of the interesting lipids and protein components that enable the bend! Alternatively, you could say that 2 different membranes fuse at the NPC.
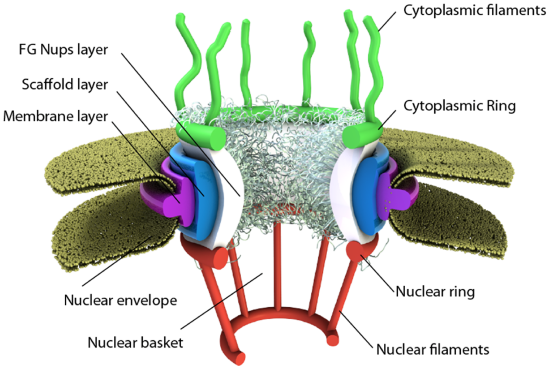
The NPC consists of 32 copies of each specific nucleoporin (Nup) except two. One of those has 48 copies and the other 16 (even these sum to 2x32 Nups). Three rings form and surround the pore. There is a 16-membered ring of Nups facing the cytoplasm (cytoplasmic ring) and another 16-membered ring of Nups facing the nucleoplasm (nuclear ring). There is 8-fold rotational symmetry in each ring, suggesting a dimeric repeat of Nups in the rings. Eight Nups in the cytoplasmic ring have a disordered end that sticks out into the cytoplasm as filaments. In contrast, the disordered ends of eight Nups in the nuclear ring form filaments which bind together to form a ring at the bottom of the nuclear basket.
The inner ring (called the FG Nups layer in the figure above), between the cytoplasmic and nucleoplasmic rings, also consists of Nups with ordered domains and disordered parts. The disordered parts of the inner ring Nups have repetitive sequences enriched in phenylalanine and glycine (hence the name FG Nups) and stick out into the central pore. These disordered region act as a filter allowing certain molecules to pass and excluding those greater with molecular weights greater than 40K. Large molecules need transport proteins called karyopherin transport factors to move through the pore.
The core structure of the NPC, obtained through cryoelectron microscopy, is shown in Figure \(\PageIndex{17\) below.
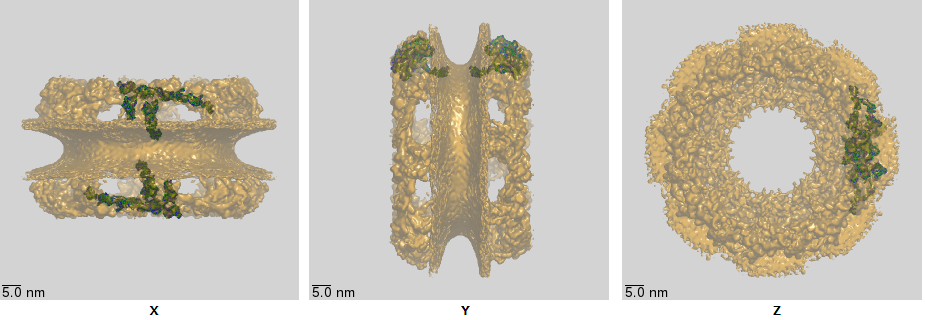
The double bilayers are very evident. The nuclear and cytoplasmic filaments, as well as the disordered FG repetitive sequences that stick into the pore from the inner ring, are not seen since they are very flexible and don't' adopt single conformations using standard structure determination methods.
Figure \(\PageIndex{18}\) shows an interactive iCn3D model of the nuclear pore complex (NPC) core (5a9q), whose structure was obtained using cryoelectron microscopy. (load slowly)
.png?revision=1&size=bestfit&width=364&height=322)
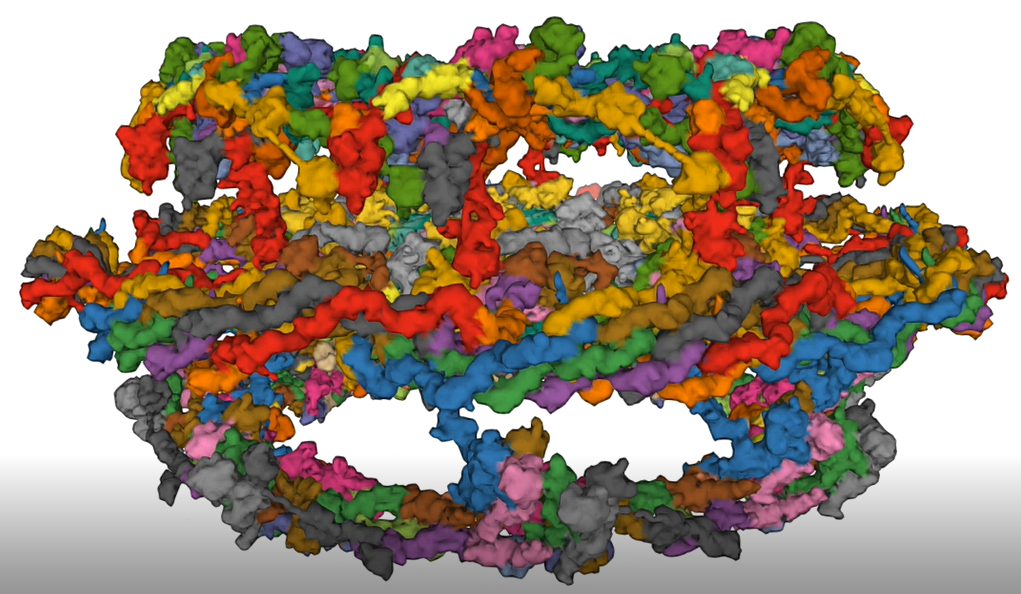
A cartoon figure showing the variety of Nups in the NPC is shown in Figure \(\PageIndex{19}\).
In this figure, the cytoplasmic and nucleoplasmic rings are both shown in green, each mostly formed by 16 copies of the Y-complex, arranged in two eight-membered rings. The inner ring, predominantly formed by 32 copies of the Nup93 complex is shown in red. Transmembrane nucleoporins are depicted in violet, and the cytoplasmic filaments and nuclear basket structure are in orange. Attached to the inner ring are Nup62 complexes (depicted in blue), which form a cohesive meshwork within the central channel through their FG-repeat domains. Not indicated is the position of Nup98, a FG-repeat-containing nucleoporin important for the transport and exclusion function of NPCs; its position in the NPC is less defined, but it might be part of the inner ring. Similarly, Aladin (also known as AAAS), Gle1, Rae1, and Npl1 (also known as hCG1 and NUPL2) have been omitted.
Large proteins and RNA that pass through the pore must first be bound to a cargo receptor, which can move the "cargo" across the pore with concomitant GTP hydrolysis. This is a process that is closer to active transport so we will discuss that in Chapter 11.3.
The entire nuclear pore complex was solved in 2022 using cryoEM. Here are two videos showing the dilated complex (7R5J). Click on the images to download mp4 animations of the complex.
 |
 |



