7.2: Polysaccharides
- Page ID
- 14956
Polysaccharides contain many monosaccharides in glycosidic links and may have many branches. They serve as either structural components or energy storage molecules. Polysaccharides consisting of single monosaccharides are homopolymers. The most common are starch, glycogen, dextran, cellulose, and chitin. We'll discuss based on whether the acetal link is alpha or beta.
α 1,4 main chain links
Starch and Glycogen: These polysaccharides are polymers of glucose linked in α 1,4 links with α 1,6 branches. Starch, found in plants, is subdivided into amylose, which has no branches, and amylopectin, which does. Starch granules consist of about 20% amylose and 80% amylopectin. Glycogen, the main CHO storage in animals, is found in muscle and liver, and consists of glucose residues in α 1,4 links with lots of α 1,6 branches (many more branches than in starch).
Here are various ways to render in 2D the chemical structure of a branched glycogen and starch fragment, as shown in Figure \(\PageIndex{1}\).
The top part of the figure shows the Haworth structure. The bottom part shows two glucose units in red and blue in the more structurally clear chair and wedge/dash representations.
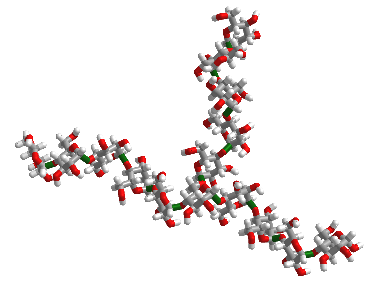
Figure \(\PageIndex{2}\) shows an interactive iCn3D model of 10 glucose monosaccharides in an α-(1,4) linkage with five glucose units with α-(1,4) linkages attached to the main chain through α-(1,6) branch at glucose 6 of the main chain. The type of substructure would be found in starch (amylopectin) and glycogen.
_linked_glucose_with_an_%25CE%25B1-(1%252C6)_branch.png?revision=1&size=bestfit&width=336&height=259) I
I
Figure \(\PageIndex{3}\) the structure in the iCn3D model in a diagrammatic fashion in which glucose is represented as a blue circle with the acetal/glycosidic/glucosidic linkages between the monosaccharides written between the circles. The 14A label shows that the acetal linkage is an α-(1,4) link with a single α-(1,6) branch.

The linkages are written in a variety of conventions. These include 14A, 14α, 4A and 4α. The between many sugars is often a 1,x link where x is 2,3, 4, 5 or 6. In those cases the 1 can be omitted. The program used to generate images in this text uses both numbers and A or B.
What makes carbohydrates so complex is their 3D structures. Like proteins and nucleic acids, they can adopt a myriad of conformations. As the monomeric units are so homogeneous, especially in homopolymers, it isn't easy to get crystal structures for them so computer models are often used.
Studies have shown that the simple starch fraction amylose α 1,4 polymer of glucose, often envisioned as a straight chain, can adopt three main conformations. They are double-helical A- (found chiefly in cereals), double helical B-(found primarily on tubers) amyloses and single-helical V-amylose (or simply A, B and V structures). The A and B do NOT represent alpha or beta in this classification system. The A and B forms consist of double helices aligned in a parallel fashion with about 6 glucoses per turn. The helices appear to be left or right-handed, and this ambiguity might arise from a lack of crystal structures.
In contrast, a well-defined structure of the V helix is known. It folds into a left-handed helix with 6 glucoses per turn and a pitch of about 8Å. Unlike alpha helices of proteins and the double-stranded helix of DNA, the center of the helix is NOT packed tightly and can accommodate small molecules. One is iodine (actually triiodide, I3-), which, when bound in amylose with a sample of starch, exhibits a dark blue color. This is the basis for starch indicators that you may have used in titration reactions in biology and chemistry courses.
In proteins, alpha helices might self-associate during folding to form a 4-helix bundle. Likewise, the helices in V-amylose can associate into bundles. Figure \(\PageIndex{4}\) shows an interactive iCn3D model of the actual structure of a V-amylose, cycloamylose 26 (1C58). It consists of a linear cycloamylose strand of 26 glucose monomers, which has collapsed to form secondary structure with 6-residue helices packed together into a tertiary structure of 4-helix bundle. The blue sphere "cartoon" color coding of each glucose residue corresponds to the blue circles in the diagrammatic representation above.
.png?revision=1&size=bestfit&width=372&height=291)
Rotate the model to explore it. Trace the chains by following the blue sphere symbolic representation for glucose as you trace the main chain. Rotate it to view down the helix axes to see the 4 holes that can each accommodate a I3-. In the menu button (=), choose Style, Chemicals, Sphere to see a spacefilling model that shows the holes within each helix. Remember, NO unoccupied holes exist within either a protein alpha helix or a double-stranded DNA molecule.
The well-known macrocyclic compound cyclodextrins (for example α-cyclodextrin) are structures equivalent to one turn of V-amylose. The V-amylose helix is partially stabilized by hydrogen bonds from donors and acceptors within the helix from the OH3 on the ith glucose and the OH2 on the ith+1 glucose as well as from the OH6 on ith glucose and OH2 on the ith + 6 glucose.
In vivo, glycogen is synthesized by the attachment of glucose monomers to a core protein called glycogenin. Figure \(\PageIndex{5}\) shows a model of a glycogen particle with glycogenin at its core.
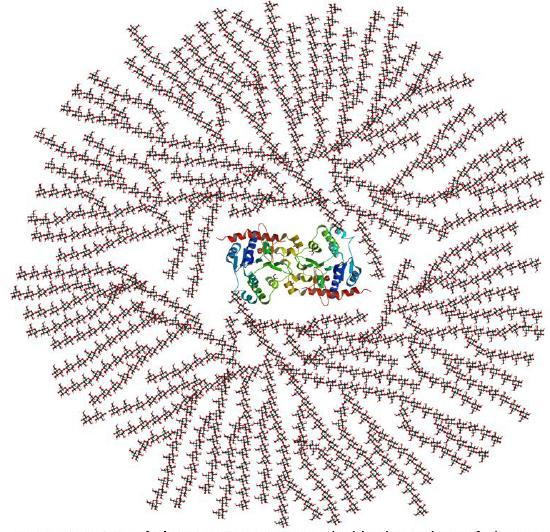
The dimeric protein glycogenin is an enzyme that autoglucosylates itself in a stepwise fashion. The first glucose is added at Tyr 195. At some point, the active site must get buried and the protein can no longer add more monomers.
It makes chemical sense to store glucose residues as either glycogen or starch, one large molecule. A review of colligative properties would inform you that if glucose was stored as the monosaccharide, a great osmotic pressure difference would be found between the outside and inside of the cell. Glycogen, with its many branchs, is a single molecule. When glucose is needed, it is cleaved one residue at a time from all the branches (at the nonreducing ends) of glycogen, producing a large amount of free glucose quickly.
Phi/Psi angles can also be used in Ramachandran plots to show the conformations around the acetal link for the starch/glycogen main chain in a fashion comparable to that for proteins (around the alpha carbon). The phi torsion angle describes rotation around the C1-O bond of the acetal link, and the psi angle describes rotation around the O-C4 bond of the same acetal link, with the glucopyranose ring considered as a rigid rotator (just as the 6 atoms in the planar peptide bond unit). The most extended form of a glucose polymer occurs when the glycosidic link is β 1,4 (as in cellulose), which forms linear chains. This would be analogous to the more extended parallel beta strand (phi/psi angles of -1190, -1130) and antiparallel beta strands (phi/psi angles of -1390, +1350) of proteins. The α 1,4 linked main chain of glycogen and starch causes the chain to turn and form a large helix. Iodine (or I3-) can fit into the helix, which turns a solution/suspension of starch blue, which turns starch purple. The less extended structure is analogous to the less extended protein alpha helix, which has phis/psi angles of -570,-470.
Figure \(\PageIndex{6}\) shows phi/psi angles for acetal/glycosidic linkage in maltose, a dissacharide of glucose, is shown below.
α 1,6 main chain links
Dextran is a branched polymer of glucose in α 1,6 links with α 1,2, α 1,3, or α 1,4 linked side chain. This polymer is used in some chromatography resins. Figure \(\PageIndex{7}\) shows chair structures (A) and wedge/dash structures (B) for dextran showing the main chain α 1,6 link with one α 1,3 branch.
Depending on its molecular weight, it is soluble in water (forming viscous solutions) and organic solvents. It is also used as a food thickener and stabilizer. It is synthesized by lactic acid-forming bacteria using sucrose as an energy source. Most uses are commercial.
β 1,4 links
Cellulose, a structural homopolymer of glucose in plants, has of β 1,4 main chain links without branching. Multiple chains are held together by intra and inter-chain H-bonds. It is the most abundant biological molecule in nature. Various rendering of 4 glucose residues in cellulose are shown in Figure \(\PageIndex{8}\). Haworth structures are not shown. Instead, more chemically informative chairs and wedge/dash structures are used. It's important to see the structures displayed in many ways, since different representations of carbohydrate structures can be found in different sources.
In A, the most common chair representation, the 2nd and 4th residues from the right-hand end are flipped versions of residues 1 and 3. Residues 1 and 2 are colored red and blue for clarity. This unit is repeated to generate the full chain. The top part of A show a simplified version of the flip of the red ring to produce the blue ring to help you see that they are indeed identical structures.
The same structure as in A is shown in the left part of B in wedge/dash from (looking down on the ring). The right-hand side of B shows a variant of the left-hand side of B that is generated by simple 1800 rotation around the bond indicated in the left of B.
In C, the simple repeat is shown without the chain flips in A and B. The acetal/glycosidic/glucosidic bond seems to be shown in a straight line in the chair structures (a bit confusing and structurally deceptive) but is shown more clearly in the adjacent wedge/dash structure.
All of the structures are correct, but the one shown in A is most often used.
One long chain of starch can interact with other chains in a structure stabilized by intrachain and interchain hydrogen bonds. Different sources display different hydrogen bonds. Some common ones are shown below. These chains align in parallel and twist to form larger cellulose fibers. Figure \(\PageIndex{9}\) shows an interactive iCn3D model of cellulose chains.

In addition, hydrophobic interactions occur between adjacent planes of cellulose strands. How can that be considering the strongly polar nature of glucose and a single cellulose strand? Figure \(\PageIndex{9B}\): below with two representations of cellulose shows how axial hydrogens projecting vertically from the plane of the glucose monomer rings form a weak but biologically significant hydrophobic surface.
Figure \(\PageIndex{9}\): Two representations showing the nonpolar axial surface of cellulose strands that contribute to hydrophobic interactions stabilizing the cellulose assemblies. The top panel shows how cellulose could interact with a nonpolar planar substance such as a layer of graphite. Axial H atoms are shown in green/cyan in the bottom panel.
Top Panel: Yang G, Luo X and Shuai L (2021) Bioinspired Cellulase-Mimetic Solid Acid Catalysts for Cellulose Hydrolysis. Front. Bioeng. Biotechnol. 9:770027. doi: 10.3389/fbioe.2021.770027. Creative Commons Attribution License (CC BY).
Bottom Panel: Uusi-Tarkka, E.-K.; Skrifvars, M.; Haapala, A. Fabricating Sustainable All-Cellulose Composites. Appl. Sci. 2021, 11, 10069. https://doi.org/10.3390/app112110069. Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/
4.0/).
The glycan is the major component in the exoskeletons of anthropoids and mollusks. It is a β 1,4 linked polymer of N-acetylglucose (GlcNAc). Compare this to cellulose which is a β 1,4 linked polymer of glucose. What a difference an N-acetyl substituent makes!
The basic chemical structure of chitin is shown in chair form in Figure \(\PageIndex{10}\) along with the symbolic nomenclature for glycans (SNFG).
Symbolic nomenclature for glycans (SNFG) -
Before we go further into the complexities of glycan structure, let's explore the symbolic nomenclature for glycan structures. The Consortium for Functional Glycomics (2005) proposed a scheme based on specific colored geometric shapes for each, as shown for the example glycan shown in Figure \(\PageIndex{11}\) for a complex glycan.
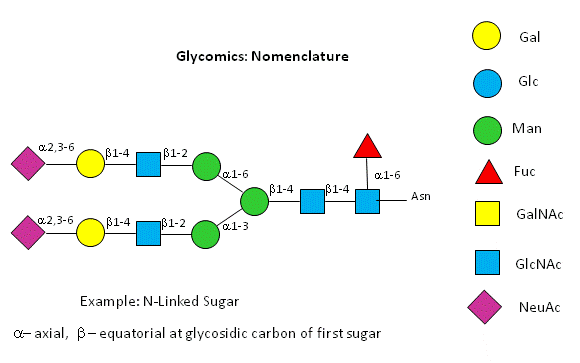
This nomenclature has recently been updated in Appendix 1B of Essentials of Glycobiology, 3rd Edition (Glycobiology 25(12): 1323-1324, 2015. doi: 10.1093/glycob/cwv091 (PMID 26543186) and is summarized in the Figure \(\PageIndex{12}\).
Glycosaminoglycans - Heteropolysaccharides with dissacharide repeating units
Many polysaccharides consist of repeating disaccharide units. A major class of polysaccharides with disaccharide repeats include the glycosaminoglycans (GAGs), all which contain one amino sugar in the repeat and in which one or both of the sugars contain negatively charged sulfate and/or carboxyl groups. The extent and position of sulfation vary widely between and within GAGs. GAGs are found in the vitreous humor of the eye and synovial fluid of joints, and in connective tissue like tendons, cartilage, etc, as well as skin. They are found in the extracellular matrix and are often covalently attached to proteins to form proteoglycans. From a bird's eye view, they are all elongated polyanions.
They and their structures are very complicated and exceedingly diverse. This makes them difficult to understand for those who want clear and unambiguous structures. From a biological perspective, they present in their local environment an incredibly diverse array of potential binding sites for ligand (both small and large). Because of these they also have functions in cell signaling. In addition, some GAGs are free-standing, others are covalently attached to proteins (a bit like glycogen is attached to glycogenin). These large molecules are called proteoglycans. We will discuss this later in Chapter 7.4 when we discuss the "carbohydrate code"
Here are the ring structures and descriptions of important GAGs. The common disaccharide repeat unit is shown twice for each structure, with the knowledge that sulfation patterns may differ for the disaccharide repeats in the actual chains. Note also that the first member of each disaccharide repeat shows the ring flipped vertically (top to bottom) as was shown in the structures for other beta-linked glycans (cellulose, chitin) above.
In a long chain, selecting which is the repeating disaccharide unit is a bit relative, as shown in Figure \(\PageIndex{13}\) for the repeating disaccharide sequence of N-acetylglucosamine (blue square) and N-acetylgalactosamine (yellow square).

In the top, the repeating units (blue-yellow) are connected to each other through beta 1,4 links while in the bottom, the connection of the repeating unit (yellow-blue) is beta 1,3. Without knowing the full chain, the best choice of annotating the repeating unit is illusive. What's most important however is to note the alternating acetal/glycosidic links throughout the whole sequence. In the figures below different disaccharide repeats are highlighted.
Hyaluronic acid
This is a polymer of glucuronate (β 1,3) GlcNAc. It offers a backbone for theattachment of protein and other GAGs. It's the only GAG without sulfate. Figure \(\PageIndex{14}\) shows a tetrasaccharide fragment with two disaccharide repeats. The internal acetal/glycosidic link of the illustrated disaccharide repeat is β 1,3 while the connection between the disaccharides is β 1,4. For one last time, the vertical flip of the glucuronic acid is shown to allow a better understanding of its flipped presentation in the actual GAG.
Hyaluronic acids are found in a variety of locations including synovial fluid, the extracellular matrix and skin, where it helps control skin moisture. It is water soluble and displays twin antiparallel left-oriented helices. Covalent conjugates of the chemotherapeutic drugs doxorubicin and camptothecin linked to hyaluronic acid, whose overall structure is similar to "worm-like micelles", have been used successfully to treat skin cancers.
Keratan sulfate
This GAG contains repeats of N-acetyl-D-glucosamine-6-phosphate in β 1,3 link to D-galactose or D-galactose-6-sulfate. The link between Gal and the modified glucosamine is β 1,4. Keratin sulfate is highest abundant in the cornea of the eye but is also found in other connective tissues such as bone, cartilage and tendon, as well as in the
central and peripheral nervous system.
Figure \(\PageIndex{15}\) shows a tetrasaccharide containing two repeating disaccharides.
Chondrotin sulfate
Th repeat dissacharide unit is D-glucuronate β(1,4) GalNAc-4 or 6-sulfate. It's found in connective tissue matrix as well as the cell surface (in the form of proteoglycans), in basement membranes, as well as intracellular granules. A tetrasacccharide showing two disaccharide repeats is shown in Figure \(\PageIndex{16}\).
Dermatan sulfate
This glycosoaminoglycan is similar to chondroitin sulfate. It is first made as a polymer of the disaccharide unit of D-gluconic and N-acetyl-D-galactosamine. The gluconic acid is epimerized to L-iduronic acid, followed by sulfation. Its structure is shown in Figure \(\PageIndex{17}\).
Heparin
This GAG contains a highly trisulfated disaccharide repeat as shown in Figure \(\PageIndex{18}\). Note that the molecule can contain glucuronate or iduronate, and the degree of sulfation of the chains varies. (Remember, there is no genetic code that specifies the actual sequence or sulfation pattern in these polymers.)
Most people are familiar with the anti-clotting properties of heparin administered as a drug. Heparin acts as a "catalyst" to accelerate the inhibition of the enzyme thrombin, which cleaves fibrinogen and activates platelets to form clots, by the blood protein antithrombin III. Heparin works in two ways to facilitate thrombin inactivation. It has a specific binding site for antithrombin III which causes a conformation in the protein, making it a more effective inhibitor. Thrombin, a positively-charged serine protease, can bind the heparin, a polyanion, nonspecifically. When it does, it diffuses along the heparin chain, where it can find bound antithrombin III much more quickly than if the inhibitor was free in the blood. Heparin effectively changes the search path of thrombin from a 3D to a 1D search.
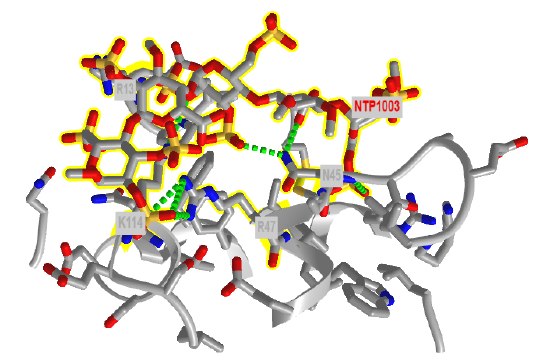
Figure \(\PageIndex{19}\) shows an interactive iCn3D model of the amino acids in antithrombin III within seven angstroms of a bound heparin 5mer (1NQ9). Dotted lines represent hydrogen bonds and salt bridges between the two. Heparin is highlighted in yellow
Agarose
Agarose is the main polysaccharide component derived from red algae. Agarose is a polymer of a disaccharide repeat of (1,3)-β-D-galactopyranose-(1,4)-3,6-anhydro-α-L-galactopyranose, is often used for a gelable solid phase for electrophoresis of nucleic acid and as a component of chromatography beads. As with starch, which is present as mixtures of amylose and amylopectin, agarose is often found with agaropectin, which is a sulfated galactan. A tetrasaccharide fragment with two dissacharide repeats is shown in Figure \(\PageIndex{20}\).



