5.7: Binding - Enzyme Linked Immunosorbant Assays (ELISAs)
- Page ID
- 83738
Introduction
Enzyme-linked immunosorbent assays (ELISAs) are used widely in biotechnology, pharmaceutical and clinical medicine labs. At the same time, they appear to be underrepresented in chemistry and biochemistry curricula, even though their sensitivity, selectivity, and ease of use would argue for their widespread use
ELISAs use primary antibodies specific to a target analyte (or antigen) as a central part of the assay. One of them is Immobilized in wells of a multiwell plate or on a strip. The species not immobilized is then added, and an immobilized analyte (antigen)-antibody complex forms. After washing the bound complex on the solid phase support to remove unbound species, a second labeled antibody is added for detection. This secondary antibody binds to the distal end (Fc domain) of the bound antibody and not the analyte. The label on the second antibody can either be a fluorophore or an enzyme that can interact with an added substrate that will generate a colored solution. The color development is then measured with a fluorometer, spectrophotometer, or if just visually if quantitation is not needed.
There are several variants of ELISAs, including the traditional ELISA, in which the antigen is bound or fixed to the surface of the solid support, or a sandwich ELISA, in which the antibody is bound to the surface. In the latter case, a second labeled antibody that binds to the antigen must bind at a different site (or epitope) on the antigen. For sandwich ELISAs, the target analyte must be large enough to accommodate two antibodies binding to different sites on the same molecule.
Cartoon diagrams showing the binding interactions in traditional and sandwich ELISAs are shown in Figure \(\PageIndex{1}\).
Figure \(\PageIndex{1}\): Binding interactions in traditional and sandwich ELISAs. Abbreviations are: fixiertes (fixed or immbolized), substrat (substrate), farblos (colorless), farbig (colored), enzymegekoppelter (enyzme coupled), antikörper (antibody), zweitantikörper (second antibody), erstantikörper (primary antibody). https://commons.wikimedia.org/wiki/File:ELISA.svg. Creative Commons Attribution 4.0 International license.
Note that both types have direct and indirect versions.
Some of the steps in a traditional ELISA are shown in Figure \(\PageIndex{2}\).
Figure \(\PageIndex{2}\): Steps in a traditional ELISA
In (1), an analyte is added to a well (outlined in gray). The solution is removed after a predefined time, and then a specific amount of analyte (such as protein) is irreversibly adsorbed (2). A blocker such as bovine serum albumin (BSA) or milk is added to bind any sites on the plate that could still bind protein. A primary anti-analyte protein is added. Some bind to the immobilized protein, and the rest is free in solution. After washing, only the primary antibody-immobilized analyte remains.
Now if a test solution (for example from a patient's blood) is added after blocking but before the addition of the primary antibody, it remains in the well unbound (5). If a primary antibody is added to this well, the immobilized antigen competes with the free antigen for binding, so less secondary antibody would be bound to the well (6). After washing, wells 4 and 7 remain. After additional rounds of washing and blocking, the secondary labeled antibody is added. Wells (such as 7) that contain patient analyte will bind less secondary antibody to the immobilized protein. When a substrate for the conjugated enzyme is added, the solution will be less colored after a defined incubation time.
In assays, a standard curve is made using a range of known concentrations of the analyte. The more analyte in the standard, the greater the competition with the immobilized analyte for the solution phase primary antibody, which would lead, after washing away the solution phase antigen:primary antibody complex, to a lower absorbance in the well with the higher solution phase antigen in the sample.
Now let's consider a sandwich assay. Instead of immobilizing a protein antigen, an antibody that binds the target antigen is immobilized. For example, an antibody against a surface SARS-Cov2 protein can be immobilized. Next, a specimen (saliva, nasal swab) containing the target surface protein recognized by the immobilized antibody is added. The great the viral load, the more SARS-Cov2 binds to the antibody. Then a second labeled antibody can be added that recognizes a different protein (the spike protein from SARS-Cov2) is added. After washing, the substrate is added, and the color development from the action of the enzyme on the substrate is measured. In this case, the more SARS-Cov2 present in the sample, the higher the signal (absorbance or fluorescence).
ELISAs have detection limits varying between 0.01 pg/mL to 100 ng/mL [1]. Although they are extensively used in health fields, they are not widely used in undergraduate biochemistry or chemistry courses, nor are they mentioned in the ACS’s Guidelines and Supplements for either Analytical Chemistry or Biochemistry. Given their importance, we choose to discuss them in this ext.
ELISA Data Analysis
The most difficult parts about ELISAs are developing an understanding of the chemical and mathematical equations, choosing/using modeling and analysis software, and evaluating validity/reliability. The typical data analysis is based on the generic Hill equation instead of the classical hyperbolic binding curve analysis we derived for a single ligand to a single site on a macromolecule. The Hill equation has more empirical parameters to use when fitting bind curves.
Here is the Hill Equation equation that we studied before.
\begin{equation}
Y=\frac{L^{n}}{K_{D}+L^{n}}
\end{equation}
For more complicated ELISA data, when a standard curve of known concentrations is used, and output signals (fluorescence, absorbance) vary from some minimum to a maximum value, the similar but more empirically useful Logistic Equation is used:
\begin{equation}
Y^{\prime}=M\left(\frac{x^{n}}{k+x^{n}}\right)
\end{equation}
where Y is the observable signal and M is the maximal observable signal. The maximal signal might not be actually observed in a ELISA assay as was the case in the binding of a ligand to a macromolecule when ligand concentrations >>KD could not be reached.
Let's use a variant of the Hill equation using the L50 value, the ligand concentration at half-maximum binding.
\begin{equation}
\begin{gathered}
0.5=\frac{L_{50}^{n}}{K_{D}+L_{50}^{n}} \\
1=\frac{2 L_{50}^{n}}{K_{D}+L_{50}{ }^{n}}
\end{gathered}
\end{equation}
hence
\begin{equation}
Y=\frac{L^{n}}{K_{D}+L^{n}}=\frac{L^{n}}{L_{50}^{n}+L^{n}}\left(\frac{\frac{1}{L^{n}}}{\frac{1}{L^{n}}}\right)=\frac{1}{\frac{L_{50}^{n}}{L^{n}}+1}=\frac{1}{\left(\frac{L_{50}}{L}\right)^{n}+1}
\end{equation}
An analogous somewhat similar equation can be derived from the logistic equation:
\begin{equation}
Y=d+\frac{a-d}{1+\left(\frac{L}{c}\right)^{b}}
\end{equation}
where four empirical parameters define the curve:
- a is the smallest measured absorbance value (blank);
- d is the largest absorbance value when Y = 1;
- c is the inflection point in the curve which can easily be seen to occur when [L]= L50= the ligand concentration at half maximal saturation; and
- b is the slope of the curve at L50 which is the Hill coefficient n. (for many ELISA curves ≈ 1).
An idealized graph of ELISA data is shown in Figure \(\PageIndex{3}\).
Figure \(\PageIndex{3}\): Idealized ELISA signal (fluorescence, absorbance) vs log [L] curve
The 4-parameter modified Logistic equation is ideal for fitting ELISA data.
An interactive active graph for the 4-parameter Logistic curve is shown in Figure \(\PageIndex{4}\). T
Figure \(\PageIndex{4}\): Interactive active graph for the 4 parameter Logistic curve.
Two of the parameters, the minimum signal a, and the maximal signal d have been set to 0.01 and 2.0 respectively, and are not changeable in the figure. Note that the greater the value of b (slope of the curve at the inflection point), the more "sigmoidal" the semilog curve appears (similar to the Hill binding curves for hemoglobin).
Lateral flow immunoassays
During the COVID pandemic, home test kits (when they were available) were used. These tests are a modified version of the sandwich ELISA described above. They differ in two main ways. The assays were not done in wells but a planar sheet in which the samples flow laterally across the sheet. As it flows across a strip through capillary action, a sample containing the SARS-COVID-2 with its spike protein would first encounter a labeled antibody to the spike protein. It would then flow into a region which contained immobilized test (anti-spike protein) and control antibodies. The bound analyte would stop and build up to sufficient concentration to see an observable colored band on the strip, but only if the sample contained the viral spike protein. These events are illustrated in Figure \(\PageIndex{5}\).
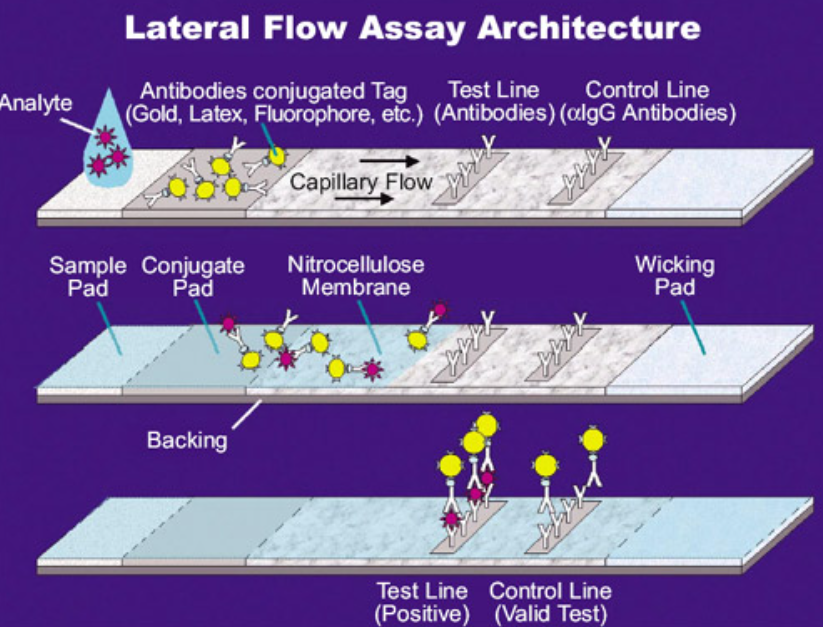
Figure \(\PageIndex{5}\): Lateral flow ELISA assay. https://en.Wikipedia.org/wiki/Latera...Flow_Assay.jpg
Figure \(\PageIndex{6}\) shows a lateral flow assay that detects the presence of serum or potentially salivary antibodies (either IgG or IgM) against the SARS-Cov2 proteins.
Figure \(\PageIndex{6}\) show a lateral flow assay which detect the presence of serum or potentially salivary antibodies (either IgG or IgM) against the SARS-Cov2 proteins. https://commons.wikimedia.org/wiki/F...0453-g002.webp. Creative Commons Attribution 4.0 International license.
The green cube represents the virus with viral proteins. It has been labeled with a gold nanoparticle (blue sphere). Gold is widely used as a labeling reagent in lateral flow immunoassays because it is chemically inert and hence extremely stable. The concentrated gold particles found in positive samples at the end of the strip can be observed visually since the gold nanoparticles absorb light through surface plasmon resonance. In this process, light of certain tunable wavelengths (based on the size of the nanoparticle) is absorbed when it matches the oscillatory frequency of the electron clouds of the metal nanoparticle. Plasmons are the oscillations in the electrons (hence electromagnetic oscillation) that occur when the nanoparticle's surface interacts with photons, causing oscillations in the electrons at the same frequency as the light (resonance).



