3.6: Chapter 3 Questions
- Page ID
- 85300
Section 1 Questions:
Q1) A small protein has the amino acid sequence below:
C1NVC2KYAPITALYC3AEEC4QQH
There are four cysteine residues in the sequence and are designated by the subscripts. The protein is digested with chymotrypsin, and the resulting slurry is followed by an anionic exchange column.
a) Where are the disulfide bonds in this structure? Are they disrupted by any of the above treatments?
b) Identify the peptide fragments created by the chymotrypsin digest. Which fragment will elute first from the anion exchange column?
A1)
a) The two disulfides are occurring between C1/C2 and C3/C4. Due to the proline in the center of this structure, we know there is likely not a folding pattern that would let another combination of disulfides to occur. No, the disulfides are not disrupted by the chymotrypsin treatment.
b) Since chymotrypsin cleaves aromatic amino acids at the C-terminus, the fragment following digestion would be:
1) C1NVC2KY
2) APITALY
3) C3AEEC4QQH
When determining which will elute off an anionic column (a positively charged bead that binds to negative residues), we need to determine the overall charge of the peptide fragments. The lysine (K) in fragment 1 will give an overall positive charge. Fragment 2 contains no charged amino acids, and the two glutamic acid (E) residues in fragment 3 will yield an overall negative charge. Since we know the column will bind most tightly to negative residues and repel positives, Fragment 1 should be eluted first, followed by fragment 2, and lastly fragment 3.
Q2) In most proteases, there is a Ser or Cys residue in the active site. Site-directed mutagenesis experiments have shown that active site Ser can be replaced with a Cys, and vice versa, with the protease, still remaining catalytically active.
a) Based on the structure of Cys and Ser, suggest an explanation as to why this could be.
b) There are other amino acids with R-groups that have similar to Ser and Cys. Hypothesize if a site-directed mutagenesis experiment changing a Ser/Cys to Thr, Tyr or Met would still retain catalytic function. Explain your reasoning.
A2) a)For both amino acids, their R-groups contain elements with unbonded lone pairs of electrons. These lone pairs on Ser (:OH) and Cys (:SH) can each act as nucleophiles and allow the protease to engage in SN-2 type reactions for proteolytic cleavage.
b) When considering the likelihood of Thr, Tyr or Met to maintain activity of the active site, the most likely candidate would be threonine. Threonine is structurally the most similar to serine and cysteine and therefore has the highest chance of not causing any steric hindrances in the active site. Threonine is however larger, so one could potentially expect a decrease in activity. Tyrosine while still containing a hydroxyl group, has a large aromatic group that would likely disrupt the interactions of the binding pocket and substrate recognition. Similarly, methionine does contain a sulfur group, but the length of the side chain most likely will cause an inhibition of substrate binding/recognition.
Q3) What pH would you use for an ion-exchange chromatography column to separate the following small peptides? Explain your answer.
Peptide 1) RGAG
Peptide 2) RGAE
Peptide 3) HGAE
Peptide 4) EGAE
A3) First, assign the formal charge on each peptide:
P1) +1
P2) Neutral
P3) -1* Note the histidine here!
P4) -2
Next to ensure that each peptide is able to maintain a unique charge, we need to assess if any pH changes could affect the net charge of the peptides and the pKas of the relevant amino acids that can contribute to the charge. R=12.48, E=4.25, H=6.00. Now, to ensure arginine is positive, glutamic acid is negative, and histidine is neutral, the pH of the column would need to be higher than 6 but less than 12.48.
Q4) Match the three letter code to the one letter code for the following amino acids.
Tyr W
Ala P
Asp A
Asn Y
Pro T
Trp D
Thr N
A4) Tyr = Y; Ala = A; Asp = D; Asn = N; Pro = P; Trp = W; Thr = T
Q5) Vasopressin is a small peptide hormone with its C-terminus converted to an amide. It is produced in the hypothamus, but ultimately becomes functionally active in the pituitary gland. One in the bioactive form, vasopressin aids in kidney function by increasing water retention, as well as increases blood pressure in the arteries. (Möller and Mari, Biochem J 2007; doi:10.1042/BJ20061480)
The peptide sequence for the mammalian protein is given below:
C1YFGNC2PRG-NH2
a) Write the sequence of vasopressin in the 3 letter code
b) Draw out the structure of the first 5 amino acids of vasopressin
c) What is the overall charge of vasopressin at pH 5? Assume the carboxylate group has a pKa of 4.0 and the amino group has a pKa of 10.
d) What is the isoelectric point of vasopressin?
A5) a) Cys-Tyr-Phe-Gly-Asn-Cys-Pro-Arg-Gly
b)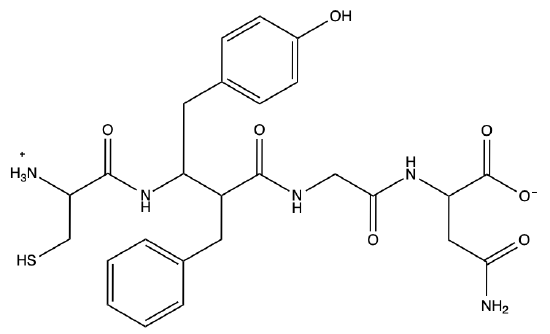
c) When we are looking at the amino acids whose R-groups can affect the overall charge of the peptide, for vasopressin that would be Tyr, and Arg. At a pH of 6 there is not enough hydroxide present to deprotonate the -OH of Tyr, so there is no charge as its pKa is 10.46. However, the pKa of Arginine is 12.48, and therefor at a pH of 6, the amine of the R-group remains protonated. Additionally, the N-terminal amino group at pH 6 contains a +1 charge and without a carboxylate group on the C-terminus to balance the charges, an additional +1 is added to the peptide. Therefore, the overall charge of the peptide is +2.
Reminder: Cys in disulfide!
d) When considering which amino acid's pKa to choose for pI calculation, you first need to determine which in the peptide can contribute to a charge on the molecule. For vasopressin, that would be Cys (pKa =8.37), Tyr (pKa = 10.46), Arg (pKa = 12.48), and the NH3+ (pKa = 10) at the N-terminus. Remember! The C-terminus of vasopressin is not a free carboxyl! Rather, it is aminated resulting in no net charge. So, we want to determine which to pKa values are the closest and straddle the pH at which the molecule has a net neutral charge.
With the pH < 8.37, the peptide has a net charge of +2. However when the pH is 10 < pH > 8.37, the cysteine residues are now deprotonated resulting in an S-. When we factor in both cysteine residue's negative charge and the +1 from the N-terminus and the +1 from arginine, the peptide is now at a net neutral!
So, to calculate the pI, we need to add the two pKa values that straddle the net neutral pH (8.37 and 10) and find the average between them. This results in a pI of 9.18!|
Section 2 Questions:
Q1) When a growing peptide chain is being synthesized by the ribosome, what terminus (amino or carboxy) is added on to? Also, which amino acid is always the "starting" amino acid for a polypeptide?
A1) Much like how DNA is always synthesized from 5' → 3', proteins are always synthesized from the amino terminus to the carboxy terminus, beginning with the start codon Methionine.
Q2) Match the type of protein structure to its definition:
a) Primary Structure (1°) 1) This structure can also be called a homodimer or heterodimer, results because of two proteins forming interactions
b) Secondary Structure (2°) 2) A growing polypeptide synthesized directly out of the ribosome
c) Tertiary Structure (3°) 3) The 3D structure of a protein, fully synthesized and correctly folded
d) Quaternary Structure (4°) 4) The protein structure formed using the R-groups of the polypeptide to create α-helices and β-sheets.
A2)
a = 2
b = 4
c = 3
d = 1
Section 3 Questions
Q1) You are considering choosing between traditional centrifugation and density centrifugation for the following scenarios. Explain which method would result in the best result for your experiment and why.
a) You want a crude cell pellet free of all supernatant.
b) From a plant cell, you want to separate the nucleus from the chloroplast.
c) You want to separate the nuclear envelope from the nucleus (hint: think about the structure of the nucleus!)
A2)
a) Traditional centrifugation. All that is needed for this experiment is the cell material to be pelleted away from the supernatant, so traditional centrifugation is sufficient.
b) Density centrifugation. To separate on an organellar level, density centrifugation should be used to increase efficiency. Bonus fun fact! Choosing an osmotically inert material such as Percoll can improve separation by not inducing hyper- or hypotonic lysis!
c) Density centrifugation. To fractionate on a suborganellar level, density centrifugation is a must. Proceeded by the appropriate experiments, an osmotic material such as sucrose can be used to fraction the nuclear envelope away from the nucleus.
Q2) You are creating a cell-free extract of Arabidopsis proteins that you want to keep for extended storage at -80°C. However, when thawed you still want the protein to remain functional for future assays and you know just adding glycerol will cause the protein to denature.
a) What is a method you can use to add glycerol to your protein extract while keeping it stable in solution? Explain why.
A2)
To increase the concentration of glycerol in the buffer without adding directly, dialysis should be used. This allows for an exchange of buffer components, ultimately bringing both solutions to equilibrium. So, if you have a higher concentration of glycerol in the dialysis solution than in the cell-free extract, dialysis will cause the concentration of glycerol in the cell-free extract to increase slowly over time, preventing protein precipiation.
Q3) The components of a cell-free extract contain: 25 mM HEPES, 100mM KCl, 5 mM MgCl2 250 mM sucrose 10% glycerol and 1 mM dithiothreitol. You dialyze with a buffer containing 25 mM HEPES, 100 mM KCl, 12 mM MgCl2, 17% glycerol, and 2 mM dithiothreitol. Hypothesize if the components of the cell-free extract will increase, decrease, or stay the same following overnight dialysis. (Li et al., 2002 doi: 10.1105/tpc.010258)
A3) The HEPES and KCl concentrations will stay the same, as both the cell extract and dialysis solution contain equal concentrations. The concentration of sucrose will decrease. The concentrations of the MgCl2, glycerol, and dithiothreitol will all increase.
Q4) Match the type of chromatography to its definition.
a) Cation-exchange 1) Antibodies are bound to beads and bind to tagged proteins
b) Affinity 2) A bead or gel matrix is created resulting in low molecular weight proteins exiting the column first, and larger last
c) Anion-exchange 3) A negatively charged bead binds to net positively charged proteins causing net negative and neutral proteins to elute faster
d) Size-exclusion 4) A positively charged bead binds to net negatively charged proteins causing net positive and neutral proteins to elute faster
e) Hydrophobic Interacting 5) A high salt concentration causes all proteins to bind to the column matrix. Decreasing the salt solution causes hydrophilic proteins to elute first, followed by hydrophobic
A4) a = 3; b = 1; c = 4; d = 2; e = 5
Q5) You want to separate 4 proteins with the following molecular weights: 120 kDa, 100 kDa, 150kDa, and 70 kDa.
a) What percentage of acrylamide gel would you use to resolve these proteins, 7%, 12%, or 15%? Explain.
b) After seeing your data from a), you decide a 25 kDa protein is also of interest. Can you use the same acrylamide % as in a)? Why or why not? And would you be able to
resolve all 5 proteins on your gel?
A5) a) A 7% gel would be the best option here. The lower percentage of acrylamide used for the gel, the larger the pores allowing for easier movement of high weight molecular weights.
b) No, a 7% acrylamide gel would not be able to resolve a 25 kDa protein. You would need a higher percentage gel, at least 12% to resolve a protein with a 25 kDa MW. With a 12% gel, it might be hard to distinguish between 100 and 120 kDa. You might consider running a gradient gel (4-15%) to see all 5 proteins.
Q6) You are studying a protein that undergoes the posttranslational modification of phosphorylation to become active, the addition of negatively charged phosphates to the outer surface of the protein. While you know the protein is phosphorylated to become active, you want to determine how many phosphates are added. When the protein is inactive (not-phosphorylated) the pI is 8.0.
What experiment could you plan to determine the number of phosphates added to the active form of your protein?
A6) You could plan to do isoelectric focusing of your protein in the inactive and active form to visualize the shift in pI. Knowing how much negative charge each phosphate adds to the pI, you can determine the number of phosphates added by how your protein moves on the isoelectric gel.
Section 5 Questions
Q1) Given the data below from a Bradford Absorbance experiment, determine the concentration of a protein extract:
Q2) The following data is from a circular dichroism experiment, based on the absorbance pattern, what is the predominant secondary structure of the protein ?
Q3) FRET Question
Q4) Question about MS with poor sequence coverage that doesn't contain a lot of basic AAs; use a new enzyme to digest.
Q5) NMR Question (use data?)
Q6) Question about Cryo-EM?



