8.8: Reverse Transcription
- Page ID
- 10446
In the central dogma, DNA codes for mRNA, which codes for protein. One known exception to the central dogma is exhibited by retroviruses. These RNA-encoded viruses have a phase in their life cycle in which their genomic RNA is converted back to DNA by a virally-encoded enzyme known as reverse transcriptase. The ability to convert RNA to DNA is a method that is desirable in the laboratory for numerous reasons. For example, converting RNAs of interest to cDNA is used in RT-PCR as well as in other applications like microarray analysis.
Process
First, one creates a DNA oligonucleotide to serve as a primer for reverse transcriptase to use on a target RNA. The primer must, of course, be complementary to a segment (near the 3’ end) of the RNA to be amplified. The RNA, reverse transcriptase, the primer, and four dNTPs are mixed. With one round of replication, the RNA is converted to a single strand of DNA. Denaturation frees the single stranded cDNA, which can be used as is, or converted to double-stranded cDNA, depending on the application.
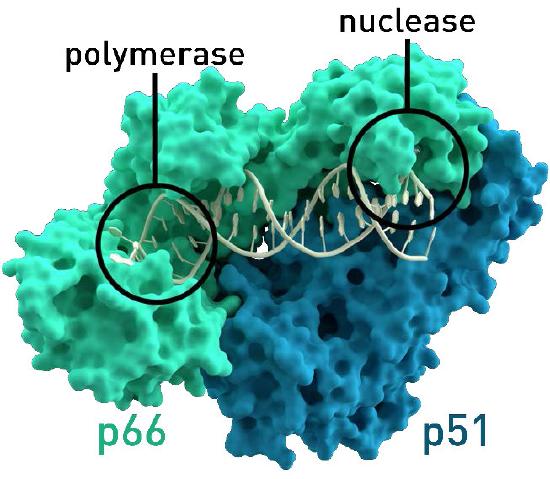
Figure 8.37 - Reverse transcriptase of HIV. The nuclease function is needed for the viral life cycle, but not for lab use. Wikipedia
Detecting molecular interactions
The study of biochemistry is basically the study of the interactions of cellular molecules. Methods for detecting interactions among biomolecules are, for this reason, very useful to biochemists. We will now discuss a couple of very different methods for detecting these inter-molecular interactions.
Yeast two-hybrid system (Y2H)

Yeast two-hybrid screening is a sophisticated technique for identifying which protein(s), out of a collection of all of a cell’s proteins, interacts with a specific protein of interest. The method relies on the interaction between two proteins to reconstitute a functional transcriptional activator within yeast cells. You may remember that many transcriptional activators are modular proteins that have a domain that binds to DNA and another domain that activates transcription (Figure 8.38).
If the transcription factor is split, so that the binding domain is attached to one protein, and the activation domain to another protein, a functional transcriptional activator can only be re-created if the two “carrier”proteins come into close proximity - that is, they interact. The presence of this functional activator can be detected by the expression of a reporter gene.
A simple way to understand this idea is by thinking of a transcriptional activator as a device, like a flashlight, that has two parts, the battery and the lamp, that must be together in order to function. Neither a person who has just a battery nor one who has only the lamp will be able to see in a dark room. But if the two interact by coming close enough to insert the battery in the flashlight, their interaction can be detected by the fact that the flashlight will now be functional as evidenced by the light produced.
It takes two to tango
Figure 8.39 (A) shows the normal yeast transcriptional activator, GAL4, with both the DNA-binding (DBD) and Activation domains (AD). It is able to stimulate transcription of the downstream reporter gene, lac z. Panels B and C show constructs that produce the GAL4 DBD and AD, respectively, fused to other proteins, one of which is termed the “bait” and the other as “prey”. Neither of these fusion proteins can stimulate transcription of the lac z gene. When constructs encoding both the bait and prey are in the same yeast cell, if the bait protein interacts with the prey, the DBD and AD of the GAL4 will be brought together to reconstitute a functional GAL4. The presence of functional GAL4 is readily detectable because it will stimulate expression of the lac z reporter gene. If the bait and prey proteins do not interact, then there will be no lac z expression. When interaction is detected through expression of the reporter gene, the specific prey protein can then be identified.
The yeast two-hybrid system allows for simultaneous screening of many prey proteins, by constructing large collections of fusion constructs, with each potential protein partner of the bait protein fused to the GAL4 activation domain.
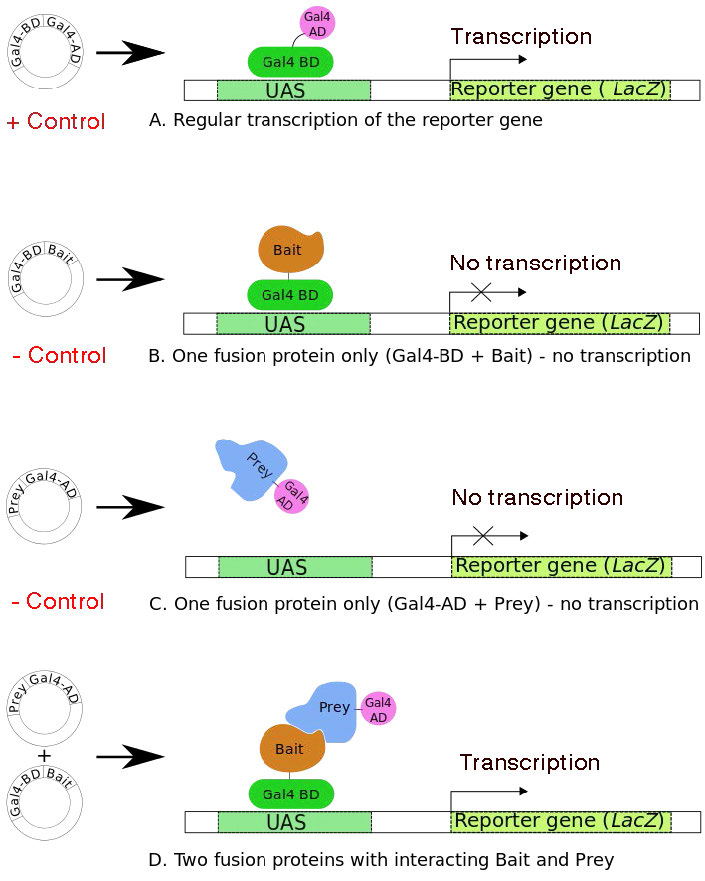
Figure 8.39 - Four scenarios for the yeast two-hybrid system. UAS = Upstream Activator Sequences - acts like a promoter. Scenario A shows that the two transcription factors start out as one protein. Wikipedia


