12.1: Introduction
- Page ID
- 16027
objectives
- Learn about the principles behind the ELISA test, and perform an experiment that simulates an ELISA test for infectious mononucleosis
- Learn another way bacteria can be identified using the Biolog microplate system (using the forehead isolate subcultured in Lab 11)
The rapid and accurate diagnosis of infectious disease is often crucial to determining patient treatment, supportive care, and precautions for the health care worker. However, some infectious diseases are more easily diagnosed than others. For example, although many types of bacteria may be easily identified by appearance on differential media, gram staining, metabolic testing, etc.… these identification methods may be time-consuming and are not always practical. In addition, not all pathogens (e.g., viruses) can be cultured in the laboratory, and so other types of diagnostic tests are necessary.
Our immune systems respond to the presence of antigens (a foreign substance that stimulates an immune response) by producing antibodies (globular proteins produced and secreted by B lymphocytes) that bind specifically to them. Many diagnostic tests take advantage of the specificity of antigen-antibody binding to detect exposure to a particular pathogen. For example, some diagnostic tests detect antibodies in a patient’s serum- their presence indicates exposure to a specific pathogen, as these antibodies will only be present if the patient has been exposed to that pathogen. Other diagnostic tests may look for specific antigens in serum (or in a cultured clinical sample) by reacting them with known antibodies in clinical laboratory tests.
In this lab, you will learn how to use a standard immunological test—an ELISA that detects serum antibodies to diagnose infectious disease. In addition, you will use the Biolog microbial identification system to identify an isolate from human skin.
ELISA TEST: The Enzyme-linked Immunosorbent Assay (ELISA) is used to detect several types of pathogens and is particularly useful for the diagnosis of viral infections. Although there are many variations of this test, it is most commonly used to detect serum antibodies to a particular viral pathogen. ELISAs are used to diagnose exposure to human immunodeficiency virus (HIV), Epstein-Barr virus (EBV) and several other pathogens. The test typically uses 96-well plates that have viral antigens bound to the wells. When serum is added, only antibodies that are specific to the antigen in the well will bind to it—all other serum components (including other types of antibodies) will be removed during washing steps. A secondary antibody (one that recognizes the first antibody) is then added. This secondary antibody is conjugated (attached) to an enzyme (horseradish peroxide is one commonly used enzyme). After washing away unbound secondary antibodies, the enzyme’s substrate is added, and produces a color reaction in the well (this is known as colorimetric detection). A color change is an indication of serum antibody, which means that the patient has been exposed to that particular pathogen. Further testing may be done to confirm a positive ELISA test.
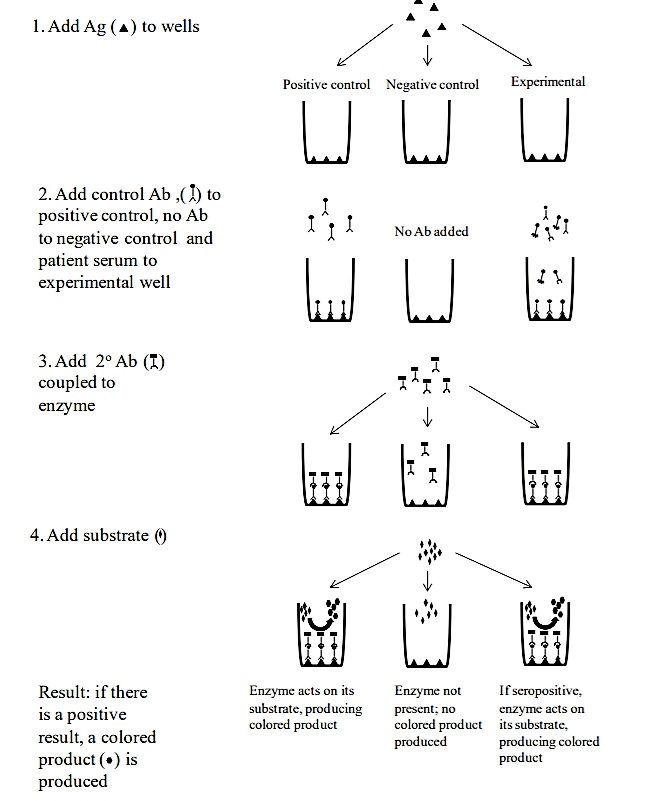.png?revision=1)
.png?revision=1&size=bestfit&width=711&height=333)
In this lab, you will be using a kit that simulates the ELISA test using non-pathogenic materials. Our simulation will show how ELISAs are used to detect infectious mononucleosis, commonly called “kissing disease” because it is transmitted by saliva. Although young children are the largest population of people affected by this viral pathogen, they generally exhibit very few symptoms. Adolescents and young adults are more likely to have symptoms of infection, including fatigue, sore throat, fever, swollen lymph nodes and, in severe cases, an enlarged spleen. The disease is caused by the Epstein-Barr virus (EBV) that is also associated with Chronic Fatigue Syndrome, and in some geographic locations, with certain types of cancers (ex: Burkitt Lymphoma)
B. Biolog System
The Biolog system is designed to identify bacteria based on their metabolic properties. This identification system uses a 96-well plate—each well contains: (a) a redox dye (tetrazolium), and (b) a substrate or chemical sensitivity test. For example, some wells contain sugars or amino acids; others test the bacteria’s ability to survive in the presence of acid, high salt concentrations, and antibiotics. There are also positive and negative control wells to ensure that the test is working correctly.
To perform the test, a suspension of bacteria is added to a 96-well microplate and incubated (usually at 33˚C). If the bacteria are able to use the substrate in a particular well, a tetrazolium dye is reduced (has electrons transferred to it), and as a result the dye in that well will turn purple. Individual species will give unique patterns of substrate utilization. These patterns can be compared to a database of known species for identification. In this experiment you will: (1) inoculate your unknown (forehead) bacterial isolate onto a Biolog microplate and next week you will (2) record the results for your unknown, and (3) use the software and database to identify your unknown bacterial species based on its pattern of substrate utilization (Steps 2 and 3 will be done in Lab 13).
Key Terms
Enzyme-linked immunosorbent assay (ELISA), antigen, antibody, colorimetric detection, secondary antibody, Epstein-Barr virus, infectious mononucleosis, Biolog, turbidimeter, tetrazolium, transmittance


