3.4: Nucleic Acids
- Page ID
- 2971
Determination of the structure of the most common form of DNA, known as the B form, was one of the most important scientific advances of the 20th century. Using data from Rosalind Franklin, James Watson and Francis Crick initiated the modern era of molecular biology with their paper in the April 25, 1953 issue of Nature. Arguably, that single page paper has had more scientific impact per word than any other research article ever published. Today, every high school biology student knows the double helical structure in which G pairs with C and A pairs with T. The DNA molecule is a polymer of nucleoside monophosphates with phosphodiester bonds between the phosphate and the 5’ end of one deoxyribose and the 3’ end of the next one. In the B form the DNA helix has a repeat of 10.5 base pairs per turn, with sugars and phosphate forming the covalent “backbone" of the molecule and the adenine, guanine, cytosine, and thymine bases oriented in the middle where they form the now familiar base-pairs that look like the rungs of a ladder.
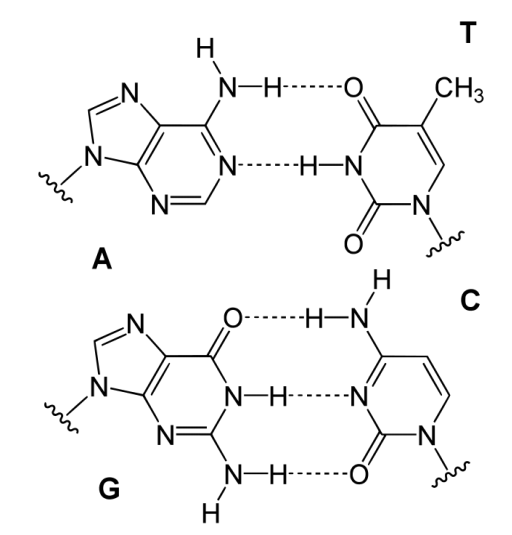
Hydrogen bonds help to hold the base pairs together, with two hydrogen bonds per A-T pair and three hydrogen bonds per G-C pair. The two strands of a DNA duplex run in opposite directions. The 5’ end of one strand is paired with the 3’ end of the other strand and vice-versa at the other end of the duplex. The B form of DNA has a prominent major groove and a minor groove tracing the path of the helix (shown at left). Proteins, such as transcription factors bind in these grooves and access the hydrogen bonds of the base pairs to “read" the sequence therein.

Other forms of DNA besides the B form are known. One of these, the ‘A’ form, was identified by Rosalind Franklin in the same issue of Nature as Watson and Crick’s paper. Though the A structure is a relatively minor form of DNA and resembles the B form, it turns out to be important in the duplex form of RNA and in RNA-DNA hybrids. Both the A form and the B form of DNA have the helix oriented in what is termed the right-handed form.
These stand in contrast to another form of DNA, known as the Z form. Z-DNA, as it is known, has the same base-pairing rules as the B and A forms, but instead has the helices twisted in the opposite direction, making a left-handed helix (Figure \(\PageIndex{3}\)). The Z form has a sort of zig-zag shape, giving to the name Z DNA. In addition, the helix is rather stretched out compared to the A and B forms. Why are there different forms of DNA. The answer relates to both superhelical tension and sequence bias. Sequence bias means that certain sequences tend to favor the “flipping" of B form DNA into other forms. Z DNA forms are favored by long stretches of alternating Gs and Cs.

Superhelicity
Short stretches of linear DNA duplexes exist in the B form and have 10.5 base pairs per turn. Double helices of DNA in the cell can vary in the number of base pairs per turn they contain. There are several reasons for this. For example, during DNA replication, strands of DNA at the site of replication get unwound at the rate of 6000 rpm by an enzyme called helicase. The effect of such local unwinding at one place in a DNA has the effect increasing the winding ahead of it. Unrelieved, such ‘tension’ in a DNA duplex can result in structural obstacles to replication.
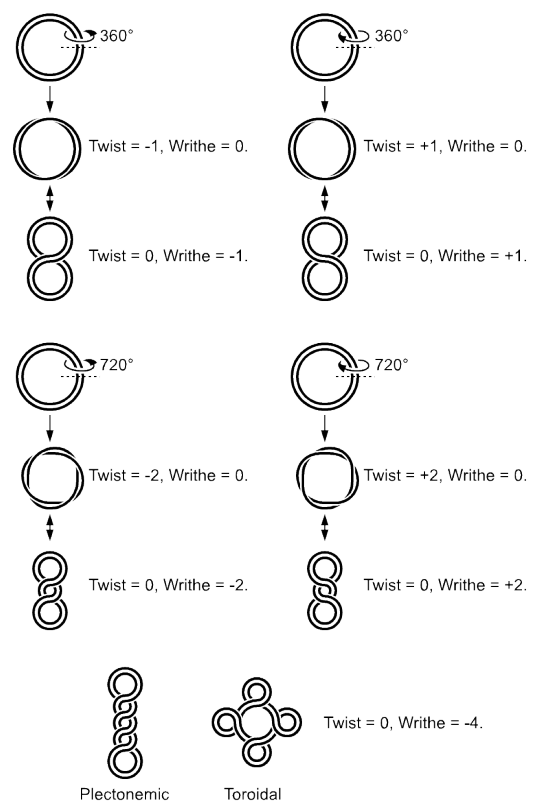
Such adjustments can occur in three ways. First, tension can provide the energy for ‘flipping’ DNA structure. Z-DNA can arise as a means of relieving the tension. Second, DNA can ‘supercoil’ to relieve the tension. In this method, the strands of the duplex can cross each other repeatedly, much like a rubber band will coil up if one holds one section in place and twists another part of it. Third, enzymes called topoisomerases can act to relieve or, in some cases, increase the tension by adding or removing twists in the DNA.
RNA Structures
With respect to structure, RNAs are more varied than their DNA cousins. Created by copying regions of DNA, cellular RNAs are synthesized as single strands, but they often have self-complementary regions leading to “fold-backs" containing duplex regions. The structure of tRNAs and rRNAs are excellent examples. The base-pairing rules of DNA are the same in RNA (with U in RNA replacing the T from DNA), but in addition, base pairing between G and U can also occur in RNA. This latter fact leads to many more possible duplex regions in RNA that can exist compared to single strands of DNA.

RNA structure, like protein structure, has importance, in some cases, for catalytic function. Like random coils in proteins that give rise to tertiary structure, single-stranded regions of RNA that link duplex regions give these molecules a tertiary structure, as well. Catalytic RNAs, called ribozymes, catalyze important cellular reactions, including the formation of peptide bonds. DNA, which is usually present in cells in strictly duplex forms (no tertiary structure, per se), is not known to be involved in catalysis.
RNA structures are important for reasons other than catalysis. The 3D arrangement of tRNAs is important for enzymes that attach amino acids to them to do so properly. Small RNAs called siRNAs found in the nucleus of cells appear to play roles in both gene regulation and in cellular defenses against viruses. The key to the mechanisms of these actions is the formation of short fold-back RNA structures that are recognized by cellular proteins and then chopped into smaller units. One strand is copied and used to base pair with specific mRNAs to prevent the synthesis of proteins from them.
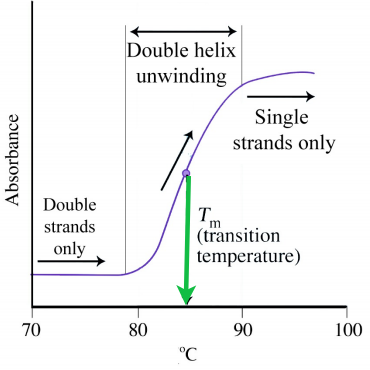
Denaturing Nucleic Acids
Like proteins, nucleic acids can be denatured. Forces holding duplexes together include hydrogen bonds between the bases of each strand that, like the hydrogen bonds in proteins, can be broken with heat or urea. (Another important stabilizing force for DNA arises from the stacking interactions between the bases in a strand.) Single strands absorb light at 260 nm more strongly than double strands (hyperchromic effect), allowing one to easily follow denaturation. For DNA, strand separation and strand hybridization are important aspects of the technique known as the polymerase chain reaction (PCR). Strand separation of DNA duplexes is accomplished in the method by heating them to boiling. Hybridization is an important aspect of the method that requires complementary single strands to “find" each other and form a duplex. Thus, DNAs (and RNAs too) can renature readily, unlike most proteins. Considerations for efficient hybridization (also called annealing) include temperature, salt concentration, strand concentration, and magnesium ion levels.